Nitrogenase 2
- Page ID
- 2508
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\) 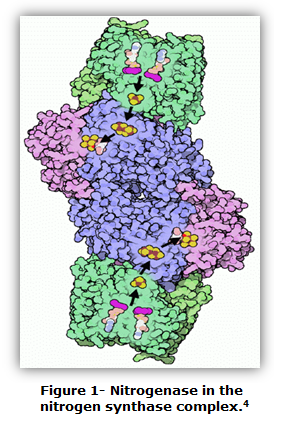
Dinitrogen gas (N2) is essential for many organisms to carry out certain roles. However, N2 that is present within organisms is not always the type needed to perform the proper functions.1 To convert N2 into a form that organisms can use, nitrogenase is often utilized. Figure 1 illustrates nitrogenase in the center of the nitrogen synthase complex. In general, nitrogenase is a metalloenzyme used to perform biological nitrogen fixation.2 Bacteria such as Azoto-bacter vinelandi and Clostridium pasteurianum utilize the metalloenzyme.3 In another example, plants need nitrogenase to obtain mineral elements from the soil. Nitrogenase, overall, reduces N2 into NH3 (ammonia).1
There are at least three different types of nitrogenase known including both Vanadium (V) nitrogenase and iron (Fe) nitrogenase2, 5 . These forms of nitrogenase are often found in bacteria.3 The commonly studied and used form of the metalloenzyme is the molybdenum (Mo) nitrogenase.2 It involves a Fe protein and MoFe protein.6 The Fe protein is composed of a [4Fe-4S] cluster and MgATP proteins that help send electrons to the MoFe protein.7 Meanwhile, the MoFe protein consists of a FeMo active site and P-cluster [8Fe-7S] metals that serve as an intermediate for transferring electrons.7 Equation (1) illustrates the complete reaction of the reduction of N2 .
N2 + 8 H+ + 8 e− + 16 ATP → 2 NH3 + H2 + 16 ADP + 16 Pi (1)
The reduction of N2 into NH3 involves magnesium salt of ATP (MgATP) and a reducing agent such Na2S2O4 (sodium dithionite in vitro) and Fe4S4 (iron-sulfur proteins in vivo).6, 8 MgATP serves in hydrolysis; meanwhile, the reducing agent plays a role in supplying electrons.1 By and large, the Fe protein reduces the MoFe protein.1-2 During reduction, an electron is transferred from the Fe protein to the MoFe protein.1-3 The MoFe protein then undergoes a reaction that involves the hydrolysis of MgATP.2, 9 As a result, energy in the form of MgATP is used to reduce N2.2, 9 A total of eight electrons in the dissociation process are needed to reduce N2.2 In addition to N2, nitrogenase also reduces other molecules such CH3NC and C2H2.6
The active site of nitrogenase is FeMoco. It is the place where dinitrogen and substrates alike are reduced, and it does not contain any amino acids.6 Figure 2 illustrates a crystal structure of the FeMoco active site, which was discovered in 1992.10 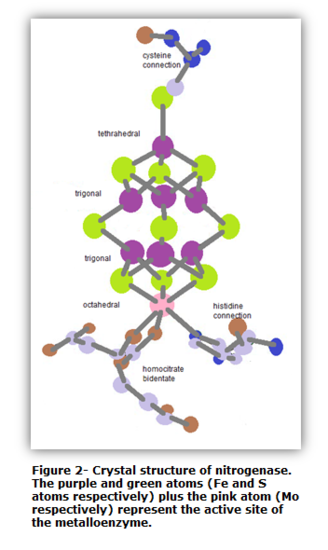
The vital feature of the FeMoco active site is the Fe7MoS9 cluster situated in the center. Located at one end of the cluster is a Mo atom, and at the other end is a Fe atom. The six-coordinate Mo atom is attached to a histidine and bidentate homcitrate chain as well as to three S atoms.10 Collectively, an octahedral structure is formed. Thus, the Mo atom with its attached branches have Oh symmetry. Meanwhile at the other end of the cluster, the Fe atom is attached to a cysteine chain and three S atoms. The Fe atom has a tetrahedral coordination overall. Thus, the Fe atom along with its substituents has Td symmetry. In the center of the cluster are six Fe atoms. Each Fe atom is attached three S atoms. In addition, each Fe atom has a trigonal geometrical structure. The point group of each Fe metal is D2d11. As a whole, the metalloenzyme is large and thus does not have a high symmetry. The overall symmetry is C1.
The structure of the active site can be used to predict whether the metalloenzyme is either IR or Raman active. In general, molecules that have a center of inversion are mutually exclusive.3 This means that IR and Raman spectra will have different frequencies. More importantly, it means that the molecule can only be either IR active or Raman active. A molecule that has a center of inversion cannot be both IR and Raman active.3 Since the active site for nitrogenase does not have a center of inversion, this suggests that the metalloenzyme may be both IR and Raman active.
EPR spectroscopy is the technique often used to study the nitrogenase and its cluster. EPR experiments show that MoFe cofactor posses a 3/2 spin state.12 Information such as this spin state give a valuable spectroscopic fingerprint that can help detect what will happen to nitrogenase under different circumstances, i.e. what happens to the active site when it is purified or isolated.12
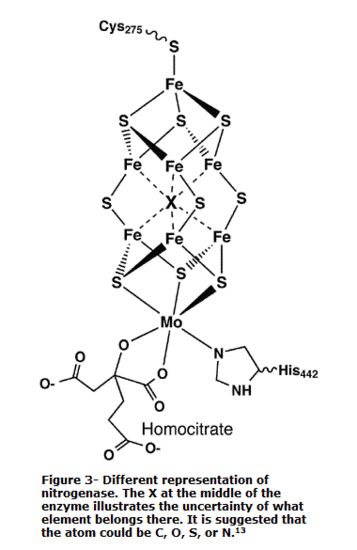 Figure 3 illustrates another and more accurate representation of the active site. The primary difference between figure 2 and figure 3 is the atom located in the center of the cluster. Figure 2 illustrates a sulfur atom in the center. Meanwhile, figure 3 illustrates an X, signifying an unknown. An interesting fact about the Fe7MoS9 cluster is that the central atom is unknown. The debate on determining the atoms continues today. However, there have been speculations that the central atom is either a S, C, O, or N atom. Different studies have strongly considered the central atom to be an S or N atom. Although the central atom is uncertain, studies have shown that the central atom (whatever it may be) plays an important role in the conversion of N2 to NH3. On the contrary, the exact mechanism of how the unknown atom helps in the reduction of N2 is yet to be known. All that is known is that the Fe-Se clusters are positioned in a way that allows the electrons to pass through the chains fast and efficiently.
Figure 3 illustrates another and more accurate representation of the active site. The primary difference between figure 2 and figure 3 is the atom located in the center of the cluster. Figure 2 illustrates a sulfur atom in the center. Meanwhile, figure 3 illustrates an X, signifying an unknown. An interesting fact about the Fe7MoS9 cluster is that the central atom is unknown. The debate on determining the atoms continues today. However, there have been speculations that the central atom is either a S, C, O, or N atom. Different studies have strongly considered the central atom to be an S or N atom. Although the central atom is uncertain, studies have shown that the central atom (whatever it may be) plays an important role in the conversion of N2 to NH3. On the contrary, the exact mechanism of how the unknown atom helps in the reduction of N2 is yet to be known. All that is known is that the Fe-Se clusters are positioned in a way that allows the electrons to pass through the chains fast and efficiently.
Overall, nitrogenase is a metalloenzyme that plays an important role in the biological processes. Nitrogenase converts dinitrogen gas into ammonia. The active site of the enzyme isFe7MoS9. The cluster has symmetries Td, Oh, and D2d. Overall, the cluster gives the enzyme C1 symmetry. Since the molecule does not have a center of inversion, this suggests that the molecule may both be IR and Raman active. Although it is known that the Fe-Se clusters in the active site aid in the reduction of N2, further research is needed to investigate exactly how the mechanism works.
References
Dennis R. Dean, J. T. B., and Limin Zheng, Nitrogenase Metalloclusters: Structures, Organizations, and Synthesis. Journal of Bacteriology 1993, 6737-6744.
- Brett M. Barney, H.-I. L., Patricia C. Dos Santos, Brian M. Hoffman, Dennis R. Dean, and Lance C. Seefeldt, Breaking the N2 triple bond: insights into the nitrogenase mechanism. The Royal Society of Chemistry 2006, ( ), 2277-2284.
- Sharpe, C. E. H. a. A. G., Inorganic Chemistry. 3 ed.; Pearson Education Limited: 2008.
- Nitrogenase. In Wikipedia, .PNG, M. c. o. n., Ed. 2002; Vol. 206 KB.
- Nitrogenase. http://www.ebi.ac.uk/intenz/spotlight.jsp?ec=1.18.6.1 (accessed May 23).
- Richards, R. L., Reactions of small molecules at transition metal sites: studies relevant to nitrogenase, an organomettalic enzyme. Coordination Chemistry Reviews 1995, 154, 83-97.
- Hamilton, T., Exploring the Role of the P-cluster in Electron Transfer in the Mo-dependent Nitrogenase. Montana State University: 2006.
- Nitrogenase. http://en.wikipedia.org/wiki/Nitrogenase (accessed April 19).
- Patricia C. Dos Santos, R. I., Hong-in Lee, Brian M. Hoffman, Lance C. Seefeldt, and Dennis R. Dean, Substrate Interactions with the Nitrogenase Active Sites. Accounts of Chemical Reserach 2005, 39 (3), 206-214.
- Dance, I., Understanding structure and reactivity of new fundamental inorganic molecules: metal sulfides, metallocarbohedrenes, and nitrogenase. Chem Chom 1998, 523-530.
- Jeremy M. Smith, R. J. L., Karl A. Pittard,; Thomas R. Cundari, G. L.-R.; Kenton R. Rodgers, a. P. L. H., Stepwise Reduction of Dinitrogen Bond Order by a
Low-Coordinate Iron Complex. Journal of the American Chemical Society 2001, 123, 9222-9223.
- Leigh, G. J., Nitrogen Fixation at the Millennium. Elsevier Science B.V.: Amsterdam, 2002.
- L. Noodleman, D. A. C., W.G. Han, T. Lovell, T. Liu, M.J. Thompson, R.A. Torres Quantum Bioinorganic Chemistry and Photochemistry. http://www.scripps.edu/news/sr/sr2004/mb04noodleman.html (accessed May 23, 2010).
(a) Modern Coordination Chemistry The Legacy of Joseph Chatt. The Royal Society Chemistry: Cambridge, 2002; (b) Weiguang Fu, T. V. M., Leonard E. Mortenson and Michael K. Johnson, Resonance Raman studies of the [4Fe-4S] to [2Fe-2S] cluster conversion
in the iron protein of nitrogenase. Federation of European Biochemical Societies 1991, 284 (2), 165-168.
- Gay Antonette Anunciacion (UCD)

