Methane Monooxygenase 3
- Page ID
- 2501
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction
Methane monoxygenase (MMO) is an enzyme responsible for the oxidation of methane to methanol according to the following equation. When oxidizing, the enzyme oxidize the substrate and adds on O2 to the atom.
CH4 + O2 + NAD(P)H + H+ -> CH3OH + NAD(P)+ + H2O (Green, et. al., 1985)
MMOs can also oxidide several other hydrocarbons, including aromatic and halogenated as well (Green, et. al., 1985). This enzyme belongs to a class called oxidoreductase, an enzyme that catalyzes an oxidation-reduction reaction.
Structure
Two types of MMOs are known- the soluble methane monooxygenanse (sMMO), whose active sites contain diiron centers, and the membrane-bound particulate MMO (pMMO), whose active sites contain copper (Takeguchi, et.al, 2000).
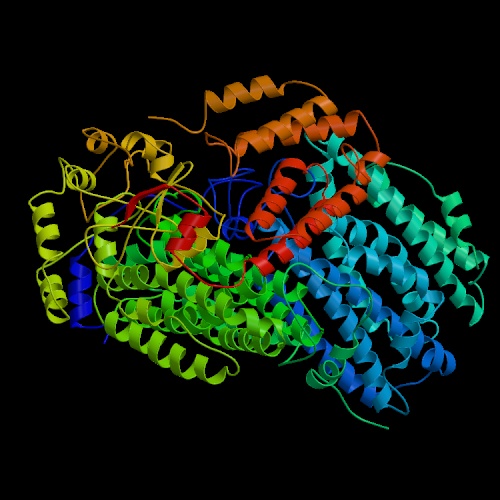
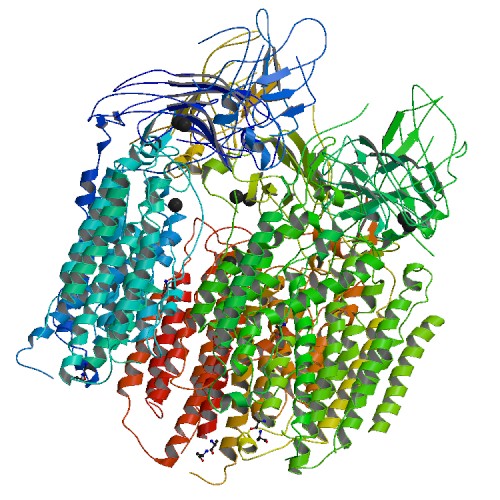
Figure 1: The ribbon structure of soluble methane oxygenase (sMMO) and particulate methane oxygenase (pMMO).
pMMO is the predominant methane oxidation catalyst in the nature; however, due to the difficulty of studying membrane-bound proteins ans the fact that pMMO is highly unstable, the mechanism of the active site of pMMO is poorly known (Takeguchi, et.al, 2000). In contrast, the soluble methane monoxygenase is the best characterized enzyme (Green, et. al.,1985). sMMO is a cytoplasmic enzyme containing a unique diiron sites at its catalytic centre (Green, et. al., 1985). It consists of 3 components: hydrolase, which is a dimer of 3 different subunits (α2, β2, and γ2), component B, and reductase.
Figure 2 below demonstrates different state of sMMO
Figure 2 : The resting, oxidized, and reduced state of the diiron core.
In the non-linear structure of sMMO, each iron bonds with six octahedral environment. The overall molecule appears to have no symmetry at all and it possesses only one C1 axis of rotation; it, therefore, belongs to the C1 point group (Housecroft, et.al., 2008).
Mechanism
Starting with MMOHred (Figure 3) , which has an FeII - FeII core, the diirons centers react with oxygen to form intermediate P, which has an FeIII - FeIII core. Compound P contains a peroxide ring where the oxygens are bound symmetrically. Next, compound P converts to the key reactive compound Q, which has a diamond-core structure. "The reaction with methane goes over the rate determining H-abstraction transition state to reach a bound-radical intermediate MMOHox , which has a bridged hydroxyl ligand interaction weakly with the methyl radical (Mogi et. al., 1999)." Finally, this intermediate rearranges intramolecularly to eliminate the alcohol and regenerate the enzyme, or MMOHred.
Figure 3: The mechanism of sMMO
References
- J. Green, H. Dalton. Protein B of Soluble Methane Monoxygenase from Methylococcus Capsulatus (Bath). J. Biol. Chem. 1985 260 (29) 15795-15801
- M. Takeguchi, I. Okura. Role of Iron and Copper in Particulate Methane Monooxzygenase of Methylosinus Trichosporium OB3b. Catalyst Surveys from Japan 4 (1) 51-63
- H. Basch, K. Mogi, D. Musaev, K. Morokuma. Mechanism of the Methane -> Methanol Conversion Reaction Catalyzed by Methane Monooxygenase: A Density Functional Study. J. Am. Chem. Soc. 1999 121 (31) 7249-7256
- C. Housecroft, A. Sharpe. Point Groups. Inorganic Chemistry. Third Edition 2008 p94
- http://en.wikipedia.org/wiki/Methane_monooxygenase

