Methane Monooxygenase 1
- Page ID
- 2499
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction and Background
Methane monooxygenase (MMO), an enzyme that belongs to the family of oxidoreductase enzymes, catalyzes the conversion of methane to methanol and is displayed in the reaction below:
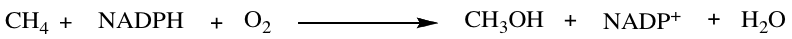
2 equivalents of NADPH split the O-O bond where one oxygen atom is reduced to water and the other is incorporated into methanol. Based on hybrid density functional (DFT) method B3LYP, the reaction is exothermic with an energy of -39 kcal/mol.
MMO enzymes serve as catalysts by converting methane into methanol at ambient temperature. Methanotrophs, organisms that utilize methane as their source of carbon and energy, possess MMO as their first metabolic pathway enzyme. Because of this property, methanotrophs are being investigated for a way to convert the most harmful greenhouse gas methane into useful sources. Commercial production of methane from natural gas is thermodynamically unfavorable because of the formation of synthesis gas, carbon dioxide and molecular hydrogen. MMO may have an impact on the use of methane as an alternative energy source because of its direct conversion of methane into methanol.
Additionally, methanotrophs can be used for bioremediation because of their ability to oxidize halogenated hydrocarbons. Methylococcus capsulatus, discovered in Bath, England, is the most studied methanotrophic bacteria.
Crystal Structure
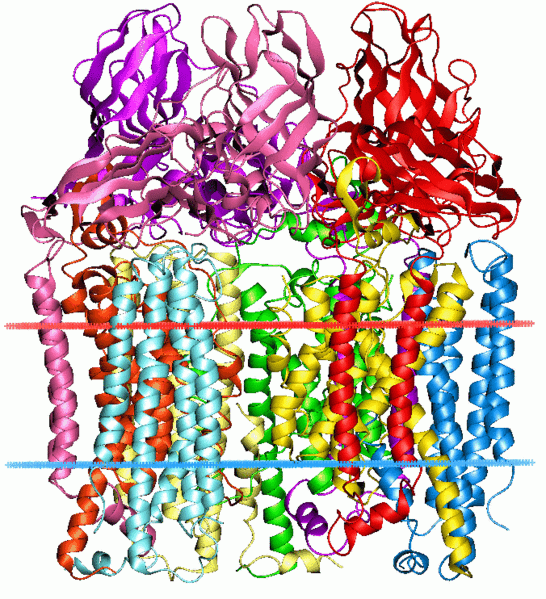
Figure 1. Crystal strucutre of methane monooxygenase
(This paragraph needs a topic sentence) 2 should be spelled out.
2 forms of MMO are known to exist: membrane bound particulate MMO (pMMO) and soluble MMO (sMMO). pMMO spans the intercytoplasmic membranes and several strains also generate sMMO under copper limiting conditions. sMMO is the most studied and contains a hydroxylase, reductase, and a regulatory protein. pMMO contains 3 subunits pmoB (α, ~47kDa), pmoA (β, ~24kDa) and pmoC (γ, ~22kDa). This report will focus on sMMO because of its availability of information.
a. Geometry of metal
Figure 2. Geometry of metal centers
(This paragraph needs a topic sentence and possibly needs to be split up into two paragraphs)
X-ray crystallography suggests that sMMO exists as a dimer, is relatively flat and has dimensions of 60 X 100 X 120 A. The 3 subunits of sMMO are α2β2γ2. Each iron atom, which is positioned in the α subunit, is dinuclear and has a coordination number of 6 making it octahedral. The nitrogen of a histidine is coordinated to both Fe1 and Fe2. The other coordinations are Fe1 to His 147, monodentate Glu 114 and Glu 144, and a water molecule. The coordination of Fe2 is similar, with two monodentate carboxylates, Glu 209 and Glu 243 and the uncoordinated oxygen atom of Glu 243 is hydrogen bound to water on Fe1, and coordination to Fe2 to His 246. As per the definition of an enzyme, the substrate must bind near the active site to catalyze the reaction. The enzyme contains hydrophobic pockets near the iron center where methane is thought to bind. Although no direct method of binding has been established, conformational changes of Phe 188 and The 213 side chains have been observed. The reduced form of MMO is achieved by a 1,3 carboxylate shift from a terminal monodentate ligand to a bridging ligand. Thus, the ligand environment becomes 5 coordinate and can now activate O2. The oxidation state of iron is now 4+ which results in a switch from low spin ferromagnetic to high spin antiferromagnetic.
Each iron atom in the MMO diiron center is octahedral by containing 6 coordination sites with 5 bound to oxygen and one bound to nitrogen. The complex is non-linear and has a C4 principle axis when the ligand groups are ignored. 4 σv planes that contain the C4 principle axis are present. Because no other symmetry elements exist, each iron center has C4v symmetry.
(This paragraph needs a topic sentence) tc
The overall point group of the enzyme is C1 because the enzymes is not linear, doesn’t have Td, Oh or Ih symmetry, doesn’t have a principle axis of rotation, has no mirror plane or center of inversion.
d. Proposed catalytic cycle
The catalytic cycles for pMMO and sMMO are proposed to be different. The catalytic cycle of copper-dependant monooxygenases, which is pMMO can be modeled after peptidylglycine α-hydroxylating monooxygenases (PHM).
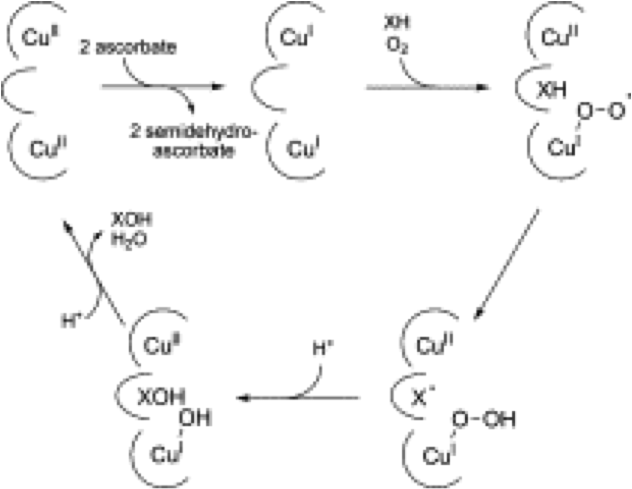
Figure 3. Catalytic cycle of pMMO
(This paragraph needs a topic sentence)TC
The intermediate formed when sMMO reacts with O2 is called “Intermediate P”. The structure is unknown but spectroscopic studies have suggested symmetrically bound oxygens. Intermediate Q is a crucial step for oxidizing MMO- this is the state where two antiferromagnetic species exist with Fe(IV) centers.
Figure 4. Catalytic cycle of sMMO
III. Symmetry
MMO is a nonlinear enzyme therefore according to the 3n-6 rule for modes where n= number of atoms in a molecule, the m=normal modes of vibration is 3(6)-6=15. According to Lippard and co-workers in the Department of Chemistry at MIT have investigated Raman active bands and have found an intermediate that is active at 800 and 900 cm-1. They have assigned this structure as being mixed valent [Fe(II) and Fe(III)].
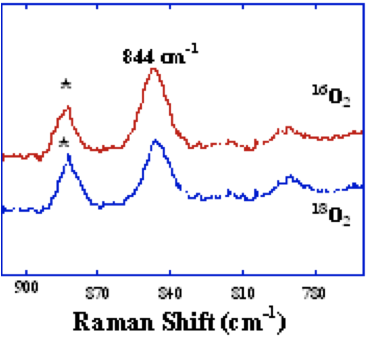
Figure 5. Raman spectrum of sMMO
References
- Rosenzweig, A. C.; Frederick, C. A.; Lippard, S. J.; Nordlund, P.; auml, Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 1993, 366 (6455), 537-543
- Lieberman, R. L.; Rosenzweig, A. C., Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 2005, 434 (7030), 177-182
- Basch, H.; Mogi, K.; Musaev, D. G.; Morokuma, K., Mechanism of the Methane‚ Methanol Conversion Reaction Catalyzed by Methane Monooxygenase: A Density Functional Study. Journal of the American Chemical Society 1999, 121 (31), 7249-7256
- Pazmiño, D.E. Torres; Winkler, M; Glieder, A; Fraaije, M.W. Monooxygenases as biocatalysts: Classification, mechanistic aspects and biotechnological applications. Journal of Biotechnology 2010, 146 (1-2), 9-24.
- http://web.mit.edu/spectroscopy/research/phys_research/lrf_05_lippard.html
- http://en.wikipedia.org/wiki/Methane_monooxygenase#cite_note-MMOcat-4

