Cytochrome P450 3
- Page ID
- 2488
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The metalloenzymes that make up the group cytochrome P-450, or CYP, play an integral and varied role in biological systems. Originally discovered in rat liver tissue by researchers in 1955, CYP has since been found in other tissues of animals, plants, fungi, and even bacteria (Lewis 1996). Understanding the functions of CYP in these biological systems unequivocally requires an understanding of CYP’s basic molecular structure and the reactivity that arises from its molecular properties.
Molecular Structure
CYP enzymes belong to a group of proteins that contain a prosthetic iron-heme cofactor. Accordingly, CYP enzymes are termed ‘hemoproteins.’ Iron holds two 4s electrons and six 3d shell electrons in its valence shell. The metalloenzyme’s reactivity depends on the shifting oxidation state of iron between ferrous Fe2+ (loss of the 4s electrons) and ferric Fe3+ (loss of an additional 3d electron) (Lewis 1996). Because the ferric state results in a half-filled 3d shell, it is the more stable form of the two states. As a result, CYP can be readily reduced with the addition of an electron.
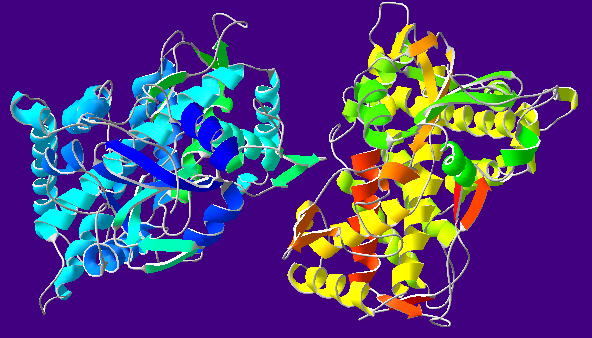
The iron center in CYP is bonded to a protoporphyrin IX macromolecule and two axial ligands. The first axial ligand is a thiolate group that is attached to the adjunct protein. The second axial ligand varies depending on the enzymatic cycle (Shaik 2005). However, in CYP’s original state, water generally occupies the axial position. Figure 1 illustrates the metal complex structure of CYP. Figure 2 shows the complete structure of a specific CYP enzyme, cytochrome P-450 Oxidase (CYP2C9).
.jpg?revision=1)
Vibrational Spectroscopy
Both Resonance Raman (RR) and Infrared (IR) spectroscopy have been used to study the structure of CYP. RR spectroscopy is particularly useful for identifying the structures of different redox and ligation states (Parthasarathi, et.al., 1987). As a result, RR has been employed to study vibrational transitions associated with the metal-ligand bonds in CYP and has provided useful insight regarding the varying oxidation and ligation states of the active site. Table 1 lists some of the distinguishing differences in the RR spectra of CYP in the Fe2+ and Fe3+ oxidation states.
| | | Frequency Range (cm-1) | |
| Mode | Symmetry | Fe(III) | Fe(II) |
| v10 | B1g | 1623-1637 | 1600-1612 |
| v37 | Eu | 1580-1601 | 1584-1586 |
| v19 | A2g | 1583 | -- |
| v2 | A1g | 1565-1584 | 1556-1564 |
| v11 | B1g | 1549-1564 | 1532-1534 |
| v38 | Eu | 1548-1550 | 1521 |
| v3 | A1g | 1485-1502 | 1462-1466 |
| v28 | B2g | 1464-1465 | 1445 |
| v4 | A1g | 1370-1373 | 1341-1344 |
Catalytic Cycle
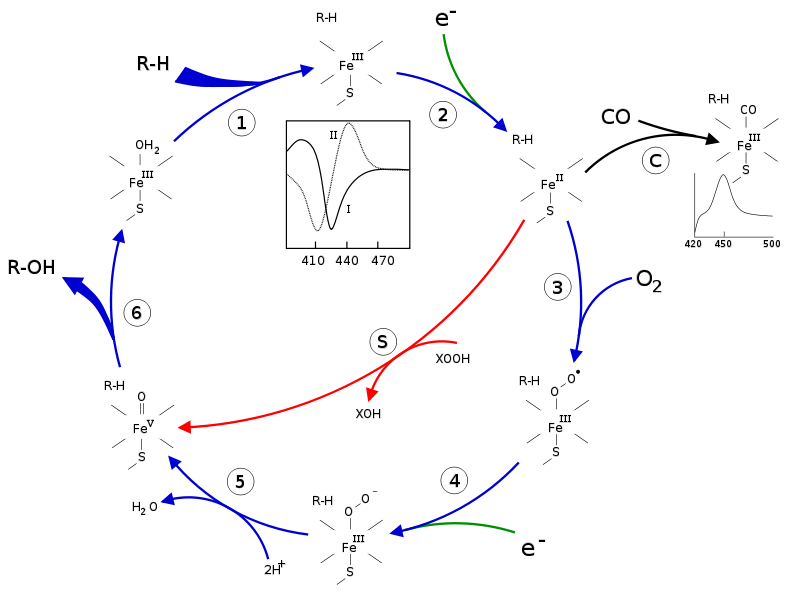
CYP can catalyze a number of reactions, including various oxidation reactions. In 1968, researchers proposed an enzymatic cycle that is commonly accepted today as the general process for CYP mono-oxygenation (Denisov, et. at., 2005). In this cycle, a substrate binds to the low-spin ferric enzyme, effectively displacing the water ligand that is coordinated to the central heme and changing the complex to a high spin-state. As a result of this shift from a low spin to a high spin state, the complex has a greater reduction potential and is more easily reduced than the original ligated heme. An oxygen molecule then binds to the heme center, forming an oxy-heme compex. The reduction of this complex, followed by two subsequent protonations and heterolysis of the O-O bond results in the formation of the original heme enzyme ligated with water and an oxygenated substrate product. Figure 3 summarized the CYP enzymatic mono-oxygenation catalytic cycle.
Biological Role
Conclusion
Biological utilization of CYP is intimately tied to CYP structure and reactivity.
References
- IMG: Cytochrome P-450 Oxidase (CYP2C9). http://en.wikipedia.org/wiki/File:Cy...idase-1OG2.png
- Degawa, M., S. Stern, M. Martin, F. Guengerich, P. Fu, K. Ilett, R. Kaderlik, F. Kadlubar. Metabolic Activation and Carcinogen-DNA Adduct Detection in Human Larynx. Cancer Research, 1994, 54, 4915-4919.
- Denisov, I.G., T. M. Makris, S. G. Sligar, I. Schlichting. Structure and Chemistry of Cytochrome P450. Chem. Rev., 2005, 105, 2253-2277
- Guengerich, F.P. Cytochrome P450s and Other Enzymes in Drug Metabolism and Toxicity. AAPS, 2006, 8 (1), 101-111.
- Lewis, D. F. V., 1996. Cytochromes P450: Structure, Function and Mechanism. Bristol: Taylor & Francis.
- Parthasarathi, N., C. Hansen, S. Yamaguchi, T. G. Spiro. Metalloporphyrin core size resonance Raman marker bands revisited: implications for the interpretation of hemoglobin photoproduct Raman frequencies. J. Am. Chem. Soc., 1987, 109 (13), 3865–3871.
- Sato, R. and T. Omura. , 1978. Cytochrome P-450. New York: Academic Press.
- Shaik, S., S. P. de Visser, 2005. Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano, P. (ed.), New York: Kluwer Academic.
- IMG: The P450 catalytic cycle. Wikipedia.org. <http://en.wikipedia.org/wiki/File:P450cycle.svg>

