5.4: Linear Regression and Calibration Curves
- Page ID
- 5728
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In a single-point external standardization we determine the value of kA by measuring the signal for a single standard containing a known concentration of analyte. Using this value of kA and the signal for our sample, we then calculate the concentration of analyte in our sample (Example 5.1). With only a single determination of kA, a quantitative analysis using a single-point external standardization is straightforward.
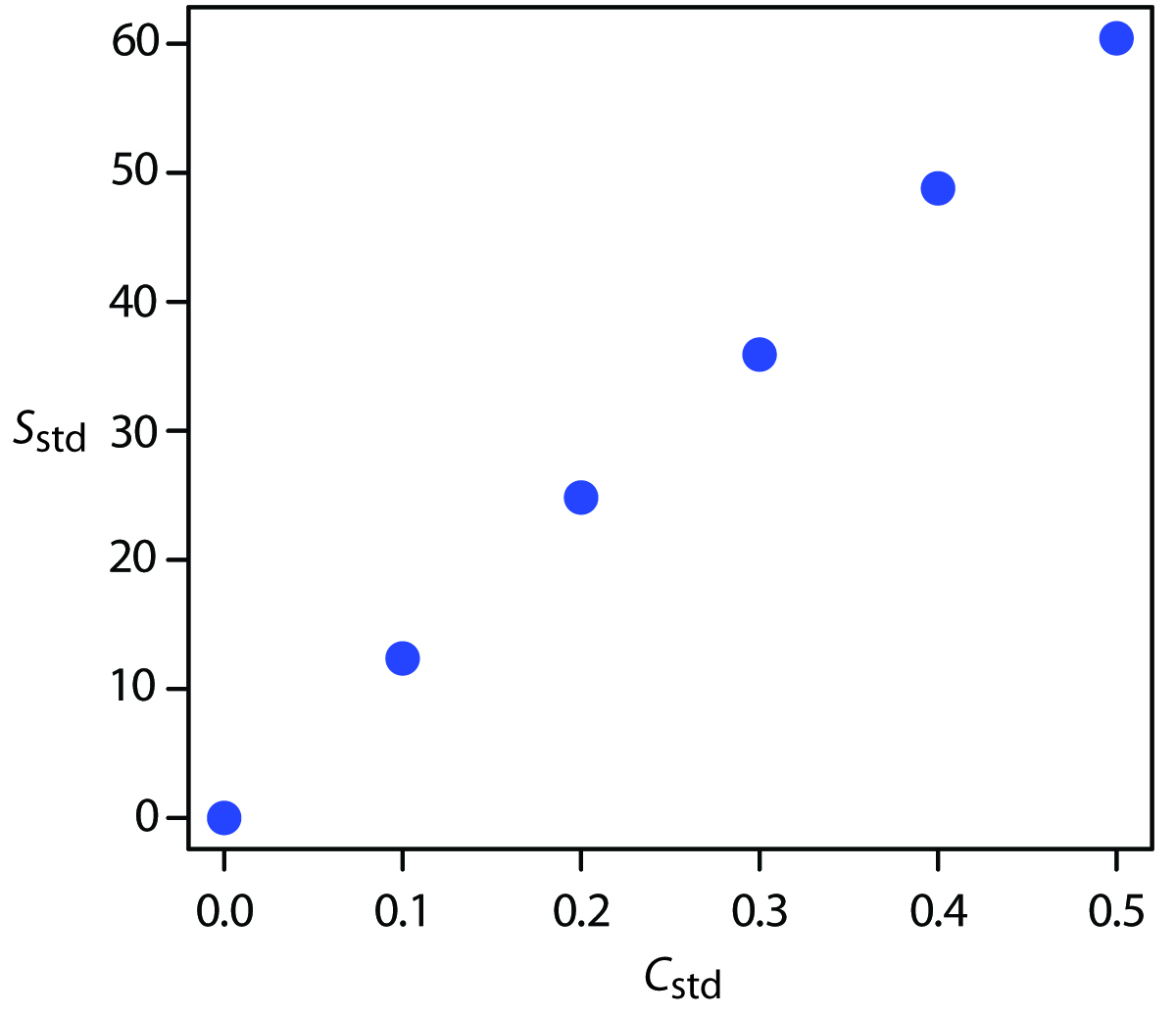
A multiple-point standardization presents a more difficult problem. Consider the data in Table 5.1 for a multiple-point external standardization. What is our best estimate of the relationship between Sstd and Cstd? It is tempting to treat this data as five separate single-point standardizations, determining kA for each standard, and reporting the mean value. Despite its simplicity, this is not an appropriate way to treat a multiple-point standardization.
| Cstd (arbitrary units) | Sstd (arbitrary units) | kA = Sstd/ Cstd |
|---|---|---|
| 0.000 | 0.00 | — |
| 0.100 | 12.36 | 123.6 |
| 0.200 | 24.83 | 124.2 |
| 0.300 | 35.91 | 119.7 |
| 0.400 | 48.79 | 122.0 |
| 0.500 | 60.42 | 122.8 |
| mean value for kA = | 122.5 |
So why is it inappropriate to calculate an average value for kA as done in Table 5.1? In a single-point standardization we assume that our reagent blank (the first row in Table 5.1) corrects for all constant sources of determinate error. If this is not the case, then the value of kA from a single-point standardization has a determinate error. Table 5.2 demonstrates how an uncorrected constant error affects our determination of kA. The first three columns show the concentration of analyte in the standards, Cstd, the signal without any source of constant error, Sstd, and the actual value of kA for five standards. As we expect, the value of kA is the same for each standard. In the fourth column we add a constant determinate error of +0.50 to the signals, (Sstd)e. The last column contains the corresponding apparent values of kA. Note that we obtain a different value of kA for each standard and that all of the apparent kA values are greater than the true value.
| Cstd | Sstd (without constant error) |
kA = Sstd/ Cstd (actual) | (Sstd)e (with constant error) |
kA = (Sstd)e/ Cstd (apparent) |
|---|---|---|---|---|
| 1.00 | 1.00 | 1.00 | 1.50 | 1.50 |
| 2.00 | 2.00 | 1.00 | 2.50 | 1.25 |
| 3.00 | 3.00 | 1.00 | 3.50 | 1.17 |
| 4.00 | 4.00 | 1.00 | 4.50 | 1.13 |
| 5.00 | 5.00 | 1.00 | 5.50 | 1.10 |
| mean kA (true) = | 1.00 | mean kA (apparent) = | 1.23 |
How do we find the best estimate for the relationship between the signal and the concentration of analyte in a multiple-point standardization? Figure 5.8 shows the data in Table 5.1 plotted as a normal calibration curve. Although the data certainly appear to fall along a straight line, the actual calibration curve is not intuitively obvious. The process of mathematically determining the best equation for the calibration curve is called linear regression.
Figure 5.8: Normal calibration curve for the hypothetical multiple-point external standardization in Table 5.1.
5.4.1 Linear Regression of Straight Line Calibration Curves
When a calibration curve is a straight-line, we represent it using the following mathematical equation
\[y=\beta_0+\beta_1x\label{5.14}\]
where y is the signal, Sstd, and x is the analyte’s concentration, Cstd. The constants β0 and β1 are, respectively, the calibration curve’s expected y-intercept and its expected slope. Because of uncertainty in our measurements, the best we can do is to estimate values for β0 and β1, which we represent as b0 and b1. The goal of a linear regression analysis is to determine the best estimates for b0 and b1. How we do this depends on the uncertainty in our measurements.
5.4.2 Unweighted Linear Regression with Errors in y
The most common approach to completing a linear regression for Equation \(\ref{5.14}\) makes three assumptions:
- that any difference between our experimental data and the calculated regression line is the result of indeterminate errors affecting y,
- that indeterminate errors affecting y are normally distributed, and
- that the indeterminate errors in y are independent of the value of x.
Because we assume that the indeterminate errors are the same for all standards, each standard contributes equally in estimating the slope and the y-intercept. For this reason the result is considered an unweighted linear regression.
The second assumption is generally true because of the central limit theorem, which we considered in Chapter 4. The validity of the two remaining assumptions is less obvious and you should evaluate them before accepting the results of a linear regression. In particular the first assumption is always suspect since there will certainly be some indeterminate errors affecting the values of x. When preparing a calibration curve, however, it is not unusual for the uncertainty in the signal, Sstd, to be significantly larger than that for the concentration of analyte in the standards Cstd. In such circumstances the first assumption is usually reasonable.
How a Linear Regression Works
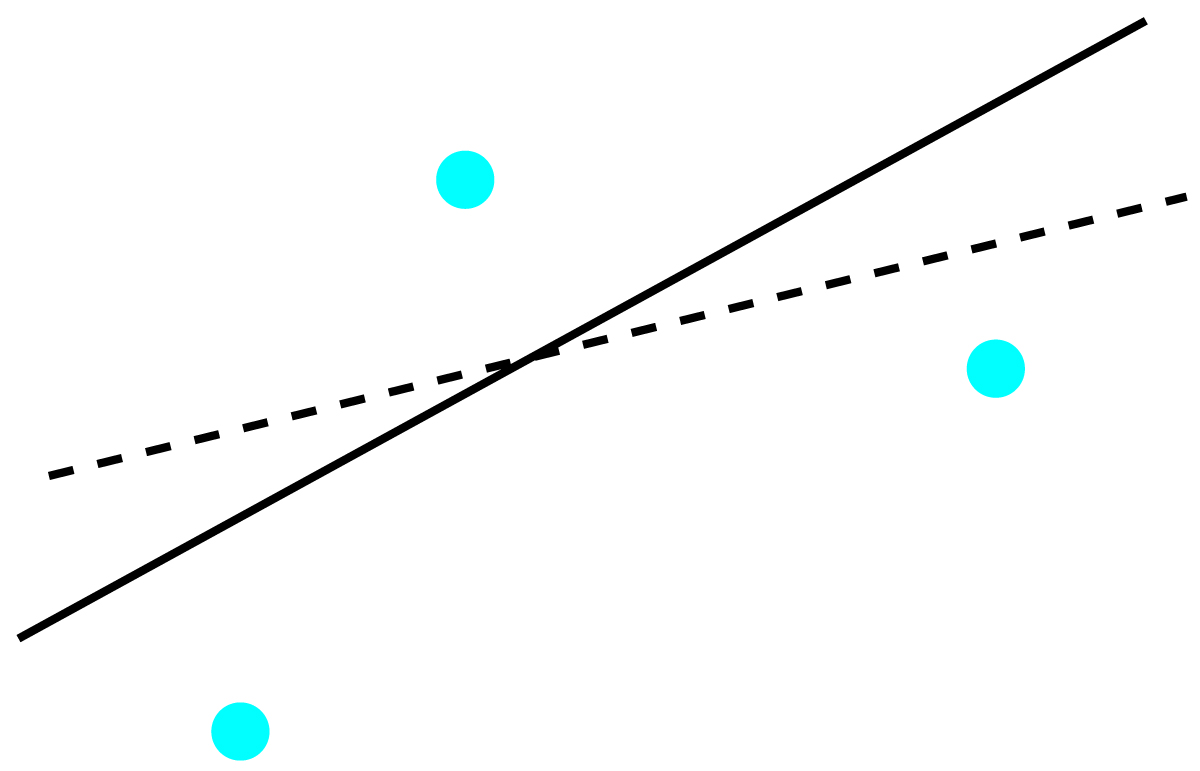
To understand the logic of an linear regression consider the example shown in Figure 5.9, which shows three data points and two possible straight-lines that might reasonably explain the data. How do we decide how well these straight-lines fits the data, and how do we determine the best straight-line?
Figure 5.9: Illustration showing three data points and two possible straight-lines that might explain the data. The goal of a linear regression is to find the mathematical model, in this case a straight-line, that best explains the data.
Let’s focus on the solid line in Figure 5.9. The equation for this line is
\[\hat{y}=b_0+b_1x\label{5.15}\]
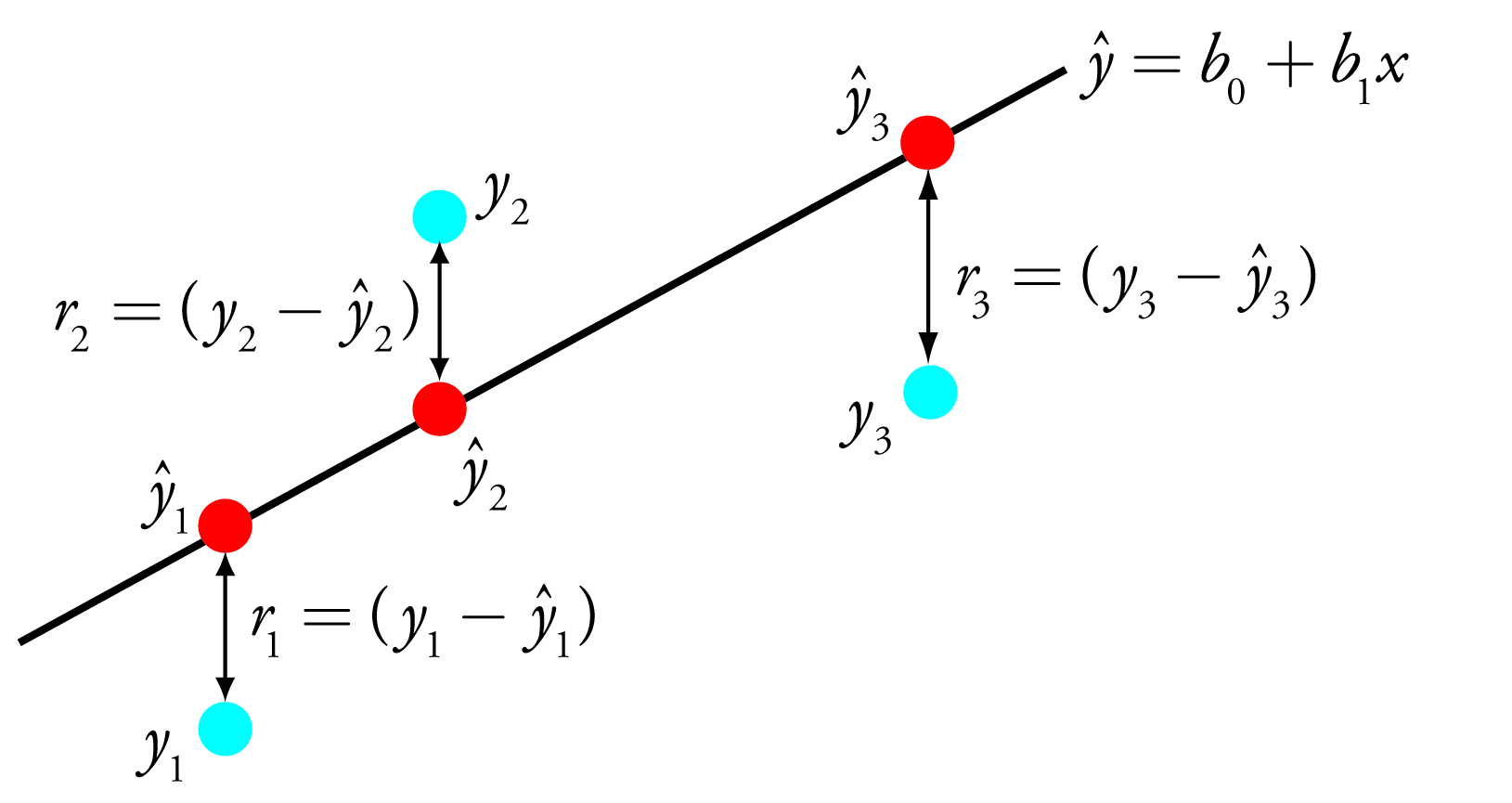
where b0 and b1 are our estimates for the y-intercept and the slope, and ŷ is our prediction for the experimental value of y for any value of x. (If you are reading this aloud, you pronounce ŷ as y-hat.) Because we assume that all uncertainty is the result of indeterminate errors affecting y, the difference between y and ŷ for each data point is the residual error, r, in the our mathematical model for a particular value of x.
\[r_i=(y_i-\hat{y}_i)\]
Figure 5.10 shows the residual errors for the three data points. The smaller the total residual error, R, which we define as
\[R=\sum_i(y_i-\hat{y}_i)^2b_1x\label{5.16}\]
the better the fit between the straight-line and the data. In a linear regression analysis, we seek values of b0 and b1 that give the smallest total residual error.
Figure 5.10: Illustration showing the evaluation of a linear regression in which we assume that all uncertainty is the result of indeterminate errors affecting y. The points in blue, yi, are the original data and the points in red, ŷi, are the predicted values from the regression equation, ŷ = b0 + b1x. The smaller the total residual error (Equation \(\ref{5.16}\)), the better the fit of the straight-line to the data
The reason for squaring the individual residual errors is to prevent positive residual error from canceling out negative residual errors. You have seen this before in the equations for the sample and population standard deviations. You also can see from this equation why a linear regression is sometimes called the method of least squares.
Finding the Slope and y-Intercept
Although we will not formally develop the mathematical equations for a linear regression analysis, you can find the derivations in many standard statistical texts.6 The resulting equation for the slope, b1, is
\[b_1=\dfrac{n\sum_ix_iy_i-\sum_ix_i\sum_iy_i}{n\sum_ix_i^2-{\sum_ix_i}^2}b_1x\label{5.17}\]
and the equation for the y-intercept, b0, is
\[b_0=\dfrac{\sum_iy_i-b_1\sum_ix_i}{n}b_1x\label{5.18}\]
Although Equations 5.17 and 5.18 appear formidable, it is only necessary to evaluate the following four summations
\[\sum_ix_i\hspace{5mm}\sum_iy_i\hspace{5mm}\sum_ix_iy_i\hspace{5mm}\sum_ix_i^2\]
Many calculators, spreadsheets, and other statistical software packages are capable of performing a linear regression analysis based on this model. To save time and to avoid tedious calculations, learn how to use one of these tools. (See Section 5.6 in this chapter for details on completing a linear regression analysis using Excel and R.) For illustrative purposes the necessary calculations are shown in detail in the following example.
Example 5.9
Using the data from Table 5.1, determine the relationship between Sstd and Cstd using an unweighted linear regression.
Hint:
Equations \(\ref{5.17}\) and \(\ref{5.18}\) are written in terms of the general variables x and y. As you work through this example, remember that x corresponds to \(C_\ce{std}\), and that y corresponds to \(S_\ce{std}\).
Solution
We begin by setting up a table to help us organize the calculation.
| xi | yi | xiyi | xi2 |
|---|---|---|---|
|
0.000 0.100 0.200 0.300 0.400 0.500 |
0.00 12.36 24.83 35.91 48.79 60.42 |
0.000 1.236 4.966 10.773 19.516 30.210 |
0.000 0.010 0.040 0.090 0.160 0.250 |
Adding the values in each column gives
\[\sum_ix_i=1.500\hspace{5mm}\sum_iy_i=182.31\hspace{5mm}\sum_ix_iy_i=66.701\hspace{5mm}\sum_ix_i^2=0.550\]
Substituting these values into Equation \(\ref{5.17}\) and Equation \(\ref{5.18}\), we find that the slope and the y-intercept are
\[b_1=\dfrac{(6\times66.701)-(1.500\times182.31)}{(6\times0.550)-(1.500)^2}=120.706\approx120.71\]
\[b_0=\dfrac{182.31-(120.706\times1.500)}{6}=0.209\approx0.21\]
The relationship between the signal and the analyte, therefore, is
\[S_\textrm{std}=120.71\times C_\textrm{std}+0.21\]
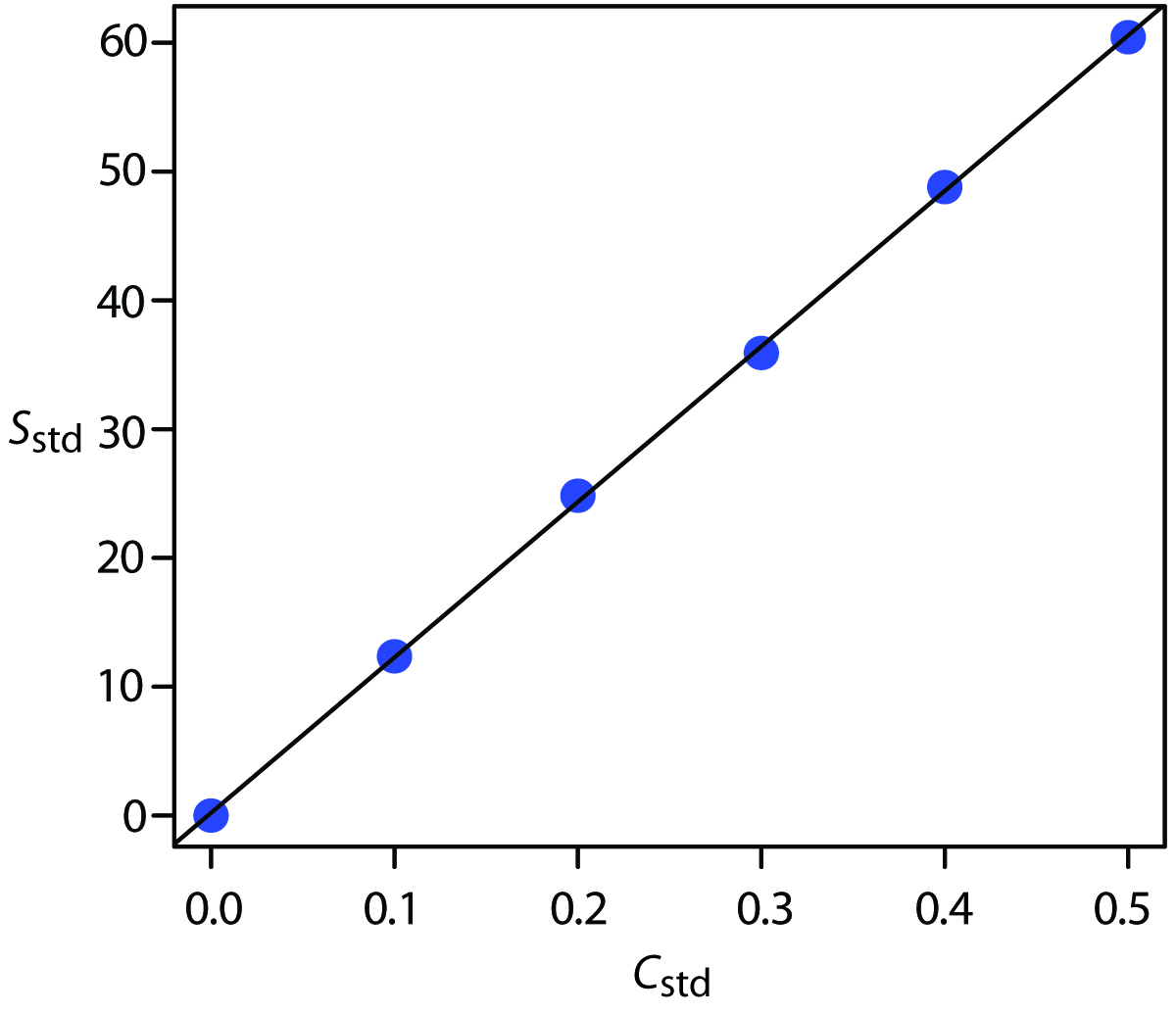
For now we keep two decimal places to match the number of decimal places in the signal. The resulting calibration curve is shown in Figure 5.11.
Uncertainty in the Regression Analysis
As shown in Figure 5.11, because of indeterminate error affecting our signal, the regression line may not pass through the exact center of each data point. The cumulative deviation of our data from the regression line—that is, the total residual error—is proportional to the uncertainty in the regression. We call this uncertainty the standard deviation about the regression, sr, which is equal to
\[s_r=\sqrt{\dfrac{\sum_i(y_i-\hat{y}_i)^2}{n-2}} \label{5.19}\]
where yi is the ith experimental value, and ŷi is the corresponding value predicted by the regression line in Equation \(\ref{5.15}\). Note that the denominator of equation 5.19 indicates that our regression analysis has n–2 degrees of freedom—we lose two degree of freedom because we use two parameters, the slope and the y-intercept, to calculate ŷi.
Did you notice the similarity between the standard deviation about the regression (Equation \(\ref{5.19}\)) and the standard deviation for a sample (equation 4.1)?
Figure 5.11: Calibration curve for the data in Table 5.1 and Example 5.9.
A more useful representation of the uncertainty in our regression is to consider the effect of indeterminate errors on the slope, b1, and the y-intercept, b0, which we express as standard deviations.
\[s_{b_1}=\sqrt{\dfrac{ns_r^2}{n\sum_ix_i^2-{\sum_ix_i}^2}}=\sqrt{\dfrac{s_r^2}{\sum_i(x_i-\bar x)^2}} \label{5.20}\]
\[s_{b_0}= \sqrt{\dfrac{s_r^2\sum_{i}x_i^2}{n\sum_{i}x_i^2 - {\sum_{i}x_i}^2}} = \sqrt{\dfrac{s_r^2\sum_{i}x_i^2}{n\sum_{i}(x_i- \overline{x})^2}}\label{5.21}\]
\[\beta_1=b_1\pm ts_{b_1}\label{5.22}\]
\[\beta_0=b_0\pm ts_{b_0}\label{5.23}\]
where we select t for a significance level of α and for n–2 degrees of freedom. Note that Equation \(\ref{5.22}\) and Equation \(\ref{5.23}\) do not contain a factor of (√n)−1because the confidence interval is based on a single regression line. Again, many calculators, spreadsheets, and computer software packages provide the standard deviations and confidence intervals for the slope and y-intercept. Example 5.10 illustrates the calculations.
You might contrast this with Equation 4.12 for the confidence interval around a sample’s mean value.
Example 5.10
Calculate the 95% confidence intervals for the slope and y-intercept from Example 5.9.
(As you work through this example, remember that x corresponds to Cstd, and that y corresponds to Sstd.)
Solution
We begin by calculating the standard deviation about the regression. To do this we must calculate the predicted signals, ŷi, using the slope and y‑intercept from Example 5.9, and the squares of the residual error, (yi - ŷi)2. Using the last standard as an example, we find that the predicted signal is
\[\hat{y}_6=b_0+b_1x_6=0.209+(120.706\times0.500)=60.562\]
and that the square of the residual error is
\[(y_i-\hat{y}_i)^2=(60.42-60.562)^2=0.2016\approx0.202\]
The following table displays the results for all six solutions.
| xi | yi | ŷi | (yi− ŷi)2 |
|---|---|---|---|
|
0.000 0.100 0.200 0.300 0.400 0.500 |
0.00 12.36 24.83 35.91 48.79 60.42 |
0.209 12.280 24.350 36.421 48.491 60.562 |
0.0437 0.0064 0.2304 0.2611 0.0894 0.0202 |
Adding together the data in the last column gives the numerator of Equation \(\ref{5.19}\) as 0.6512. The standard deviation about the regression, therefore, is
\[s_r= \sqrt{\dfrac{0.6512}{6- 2}} = 0.4035\]
Next we calculate the standard deviations for the slope and the y-intercept using Equation \(\ref{5.20}\) and Equation \(\ref{5.21}\). The values for the summation terms are from in Example 5.9.
\[s_{b_1} = \sqrt{\dfrac{ns_r^2}{n\sum_{i}x_i^2 - {\sum_{i}x_i}^2}} = \sqrt{\dfrac{6×(0.4035)^2} {(6×0.550) - (1.550)^2}} = 0.965\]
\[s_{b_0} = \sqrt{\dfrac{s_r^2\sum_{i}x_i^2}{n\sum_{i}x_i^2 - {\sum_{i}x_i}^2}} = \sqrt{\dfrac{(0.4035)^2 × 0.550}{(6×0.550) - (1.550)^2}}\]
Finally, the 95% confidence intervals (α = 0.05, 4 degrees of freedom) for the slope and y-intercept are
\[\beta_1=b_1\pm ts_{b_1}=120.706\pm(2.78\times0.965)=120.7\pm2.7\]
\[\beta_0=b_0\pm ts_{b_0}=0.209\pm(2.78\times0.292)=0.2\pm0.8\]
(You can find values for t in Appendix 4.)
The standard deviation about the regression, sr, suggests that the signal, Sstd, is precise to one decimal place. For this reason we report the slope and the y-intercept to a single decimal place.
Minimizing Uncertainty in Calibration Curves
To minimize the uncertainty in a calibration curve’s slope and y-intercept, you should evenly space your standards over a wide range of analyte concentrations. A close examination of Equation \(\ref{5.20}\) and Equation \(\ref{5.21}\) will help you appreciate why this is true. The denominators of both equations include the term \(Σ(x_i −\bar{x})^2\). The larger the value of this term—which you accomplish by increasing the range of x around its mean value—the smaller the standard deviations in the slope and the y-intercept. Furthermore, to minimize the uncertainty in the y‑intercept, it also helps to decrease the value of the term Σxi in Equation \(\ref{5.21}\), which you accomplish by including standards for lower concentrations of the analyte.
Obtaining the Analyte’s Concentration From a Regression Equation
Once we have our regression equation, it is easy to determine the concentration of analyte in a sample. When using a normal calibration curve, for example, we measure the signal for our sample, Ssamp, and calculate the analyte’s concentration, CA, using the regression equation.
\[C_\textrm A=\dfrac{S_\textrm{samp}-b_0}{b_1}\label{5.24}\]
What is less obvious is how to report a confidence interval for CA that expresses the uncertainty in our analysis. To calculate a confidence interval we need to know the standard deviation in the analyte’s concentration, sCA, which is given by the following equation
\[s_{C_\textrm A}=\dfrac{s_r}{b_1}\sqrt{\dfrac{1}{m}+\dfrac{1}{n}+\dfrac{(\bar S_\textrm{samp}-\bar S_\textrm{std})^2}{(b_1)^2\sum_i(C_{\large{\textrm{std}_i}}-\bar C_\textrm{std})^2}}\label{5.25}\]
where m is the number of replicate used to establish the sample’s average signal (Ssamp), n is the number of calibration standards, Sstd is the average signal for the calibration standards, and Cstdi and Cstd are the individual and mean concentrations for the calibration standards.7 Knowing the value of sCA , the confidence interval for the analyte’s concentration is
\[\mu_{C_\textrm A}=C_\textrm A\pm ts_{C_\textrm A}\]
where µCA is the expected value of CA in the absence of determinate errors, and with the value of t based on the desired level of confidence and n–2 degrees of freedom.
Note
Equation \(\ref{5.25}\) is written in terms of a calibration experiment. A more general form of the equation, written in terms of x and y, is given here.
\[s_x=\dfrac{s_r}{b_1}\sqrt{\dfrac{1}{m}+\dfrac{1}{n}+\dfrac{(\bar Y-\bar y)^2}{(b_1)^2 \sum_i(x_i-\bar x)^2}}\]
A close examination of Equation \(\ref{5.25}\) should convince you that the uncertainty in CA is smallest when the sample’s average signal, Ssamp, is equal to the average signal for the standards, Sstd. When practical, you should plan your calibration curve so that Ssamp falls in the middle of the calibration curve.
Example 5.11
Three replicate analyses for a sample containing an unknown concentration of analyte, yield values for Ssamp of 29.32, 29.16 and 29.51. Using the results from Example 5.9 and Example 5.10, determine the analyte’s concentration, CA, and its 95% confidence interval.
Solution
The average signal, Ssamp, is 29.33, which, using Equation \(\ref{5.24}\) and the slope and the y-intercept from Example 5.9, gives the analyte’s concentration as
\[C_\ce{A}=\dfrac{\bar S_\textrm{samp}-b_0}{b_1}=\dfrac{29.33-0.209}{120.706}=0.241 \]
To calculate the standard deviation for the analyte’s concentration we must determine the values for Sstd and Σ(Cstdi − Cstd)2. The former is just the average signal for the calibration standards, which, using the data in Table 5.1, is 30.385. Calculating Σ(Cstdi − Cstd)2 looks formidable, but we can simplify its calculation by recognizing that this sum of squares term is the numerator in a standard deviation equation; thus,
\[\sum(C_{\large{\textrm{std}_i}}-\bar C_{\large{\textrm{std}}})^2=(s_{\large{C_\textrm{std}}})^2\times(n-1)\]
where sCstd is the standard deviation for the concentration of analyte in the calibration standards. Using the data in Table 5.1 we find that sCstd is 0.1871 and
\[\sum(C_{\large{\textrm{std}_i}}-\bar C_{\large{\textrm{std}}})^2=(0.1871)^2\times(6-1)=0.175\]
Substituting known values into Equation \(\ref{5.25}\) gives
\[s_{C_\textrm A}=\dfrac{0.4035}{120.706}\sqrt{\dfrac{1}{3}+\dfrac{1}{6}+\dfrac{(29.33- 30.385)^2}{(120.706)^2\times0.175}=0.0024}\]
Finally, the 95% confidence interval for 4 degrees of freedom is
\[\mu_{C_\textrm A}=C_\textrm A\pm ts_{C_\textrm A}=0.241\pm(2.78\times0.0024)=0.241\pm0.007\]
You can find values for t in Appendix 4.
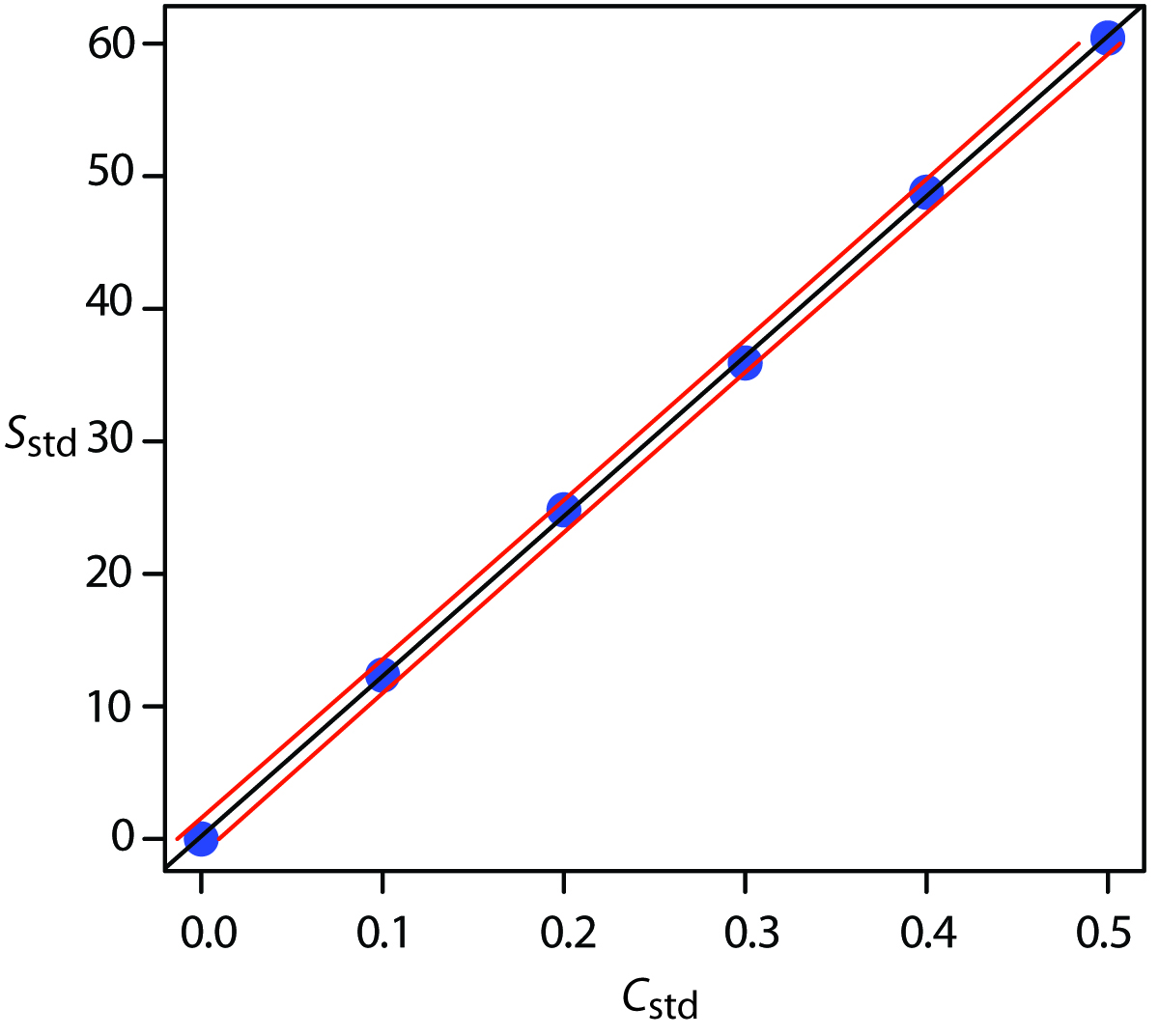
Figure 5.12 shows the calibration curve with curves showing the 95% confidence interval for CA.
Figure 5.12: Example of a normal calibration curve with a superimposed confidence interval for the analyte’s concentration. The points in blue are the original data from Table 5.1. The black line is the normal calibration curve as determined in Example 5.9. The red lines show the 95% confidence interval for CA assuming a single determination of Ssamp.
Exercise 5.4
Figure 5.3 shows a normal calibration curve for the quantitative analysis of Cu2+. The data for the calibration curve are shown here
| [Cu2+] (M) | Absorbance |
|---|---|
|
0 1.55×10–3 3.16×10–3 4.74×10–3 6.34×10–3 7.92×10–3 |
0 0.050 0.093 0.143 0.188 0.236 |
Click here to review your answer to this exercise.
In a standard addition we determine the analyte’s concentration by extrapolating the calibration curve to the x-intercept. In this case the value of CA is
\[C_\ce{A} = x\textrm{-intercept} =\dfrac{-b_0}{b_1}\]
and the standard deviation in CA is
\[s_{C_\ce{A}} = \dfrac{s_r}{b_1} \sqrt{\dfrac{1}{n} + \dfrac{(\overline{S}_\ce{std})^2}{(b_1)^2 \sum_{i} (C_{\ce{std}_i} - \overline{C}_\ce{std})^2}}\]
where n is the number of standard additions (including the sample with no added standard), and Sstd is the average signal for the n standards. Because we determine the analyte’s concentration by extrapolation, rather than by interpolation, sCA for the method of standard additions generally is larger than for a normal calibration curve.
Evaluating a Linear Regression Model
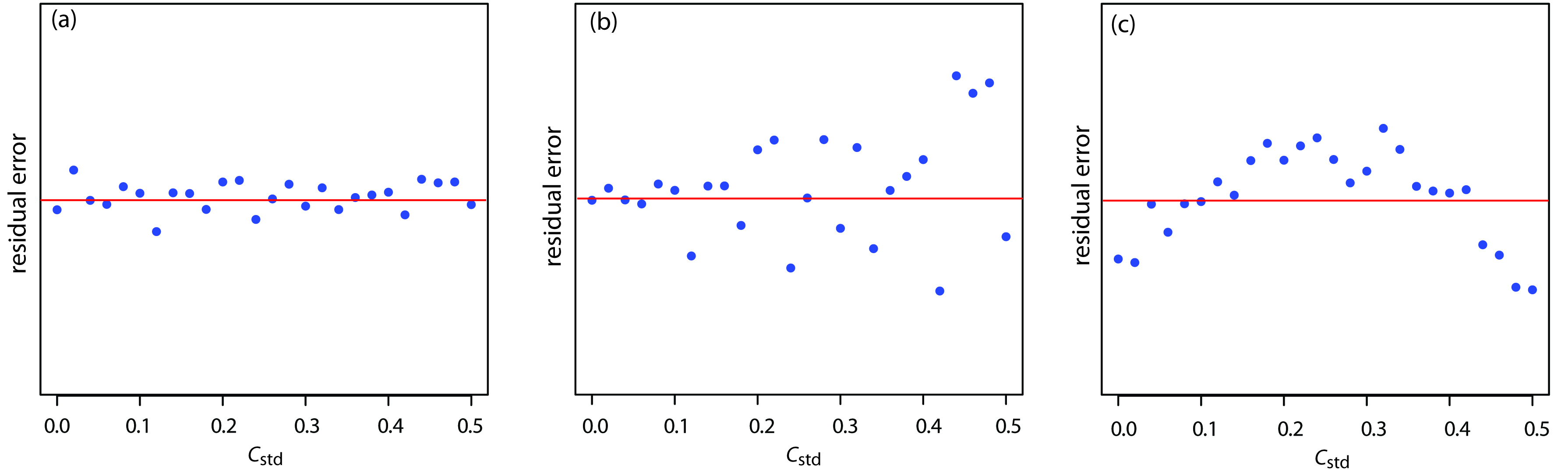
You should never accept the result of a linear regression analysis without evaluating the validity of the your model. Perhaps the simplest way to evaluate a regression analysis is to examine the residual errors. As we saw earlier, the residual error for a single calibration standard, ri, is
\[r_i = (y_i − \hat{y}_i)\]
If your regression model is valid, then the residual errors should be randomly distributed about an average residual error of zero, with no apparent trend toward either smaller or larger residual errors (Figure 5.13a). Trends such as those shown in Figure 5.13b and Figure 5.13c provide evidence that at least one of the model’s assumptions is incorrect. For example, a trend toward larger residual errors at higher concentrations, as shown in Figure 5.13b, suggests that the indeterminate errors affecting the signal are not independent of the analyte’s concentration. In Figure 5.13c, the residual errors are not random, suggesting that the data can not be modeled with a straight-line relationship. Regression methods for these two cases are discussed in the following sections.
Figure 5.13: Plot of the residual error in the signal, Sstd, as a function of the concentration of analyte, Cstd for an unweighted straight-line regression model. The red line shows a residual error of zero. The distribution of the residual error in (a) indicates that the unweighted linear regression model is appropriate. The increase in the residual errors in (b) for higher concentrations of analyte, suggest that a weighted straight-line regression is more appropriate. For (c), the curved pattern to the residuals suggests that a straight-line model is inappropriate; linear regression using a quadratic model might produce a better fit.
Exercise 5.5
Using your results from Practice Exercise 5.4, construct a residual plot and explain its significance.
Click here to review your answer to this exercise.
5.4.3 Weighted Linear Regression with Errors in y
Our treatment of linear regression to this point assumes that indeterminate errors affecting y are independent of the value of x. If this assumption is false, as is the case for the data in Figure 5.13b, then we must include the variance for each value of y into our determination of the y-intercept, bo, and the slope, b1; thus
\[b_0= \dfrac{\sum_i w_iy_i − b_1 \sum_i w_ix_i}{n} \label{5.26}\]
\[ b_1 = \dfrac{n\sum_iw_ix_iy_i − \sum_i w_ix_i \sum_iw_iy_i}{n\sum_iw_ix_i^2− (\sum_iw_ix_i)^2} \label{5.27}\]
where wi is a weighting factor that accounts for the variance in yi
\[w_i= \dfrac{n(s_{y_i})^{-2}}{\sum_{i}(s_{y_i})^{-2}}\label{5.28}\]
and syi is the standard deviation for yi. In a weighted linear regression, eachxy-pair’s contribution to the regression line is inversely proportional to the precision of yi—that is, the more precise the value of y, the greater its contribution to the regression.
Example 5.12
Shown here are data for an external standardization in which sstd is the standard deviation for three replicate determination of the signal.
|
Cstd (arbitrary units) |
Sstd (arbitrary units) |
sstd |
|---|---|---|
|
0.000 |
0.00 |
0.02 |
|
0.100 |
12.36 |
0.02 |
|
0.200 |
24.83 |
0.07 |
|
0.300 |
35.91 |
0.13 |
|
0.400 |
48.79 |
0.22 |
|
0.500 |
60.42 |
0.33 |
Determine the calibration curve’s equation using a weighted linear regression.
Hint:
This is the same data used in Example 5.9 with additional information about the standard deviations in the signal. As you work through this example, remember that x corresponds to Cstd, and that y corresponds to Sstd.
Solution
We begin by setting up a table to aid in calculating the weighting factors.
| xi | yi | syi | (syi)-2 | wi |
|---|---|---|---|---|
|
0.000 0.100 0.200 0.300 0.400 0.500 |
0.00 12.36 24.83 35.91 48.79 60.42 |
0.02 0.02 0.07 0.13 0.22 0.33 |
2500.00 2500.00 204.08 59.17 20.66 9.18 |
2.8339 2.8339 0.2313 0.0671 0.0234 0.0104 |
Adding together the values in the forth column gives
\[\sum_{i}(s_{y_i})^2 = 5293.09\]
which we use to calculate the individual weights in the last column. After calculating the individual weights, we use a second table to aid in calculating the four summation terms in Equation \(\ref{5.26}\) and Equation \(\ref{5.27}\).
| xi | yi | wi | wixi | wi yi | wixi2 |
wixiyi |
|---|---|---|---|---|---|---|
|
0.000 |
0.00 |
2.8339 |
0.0000 |
0.0000 |
0.0000 |
0.0000 |
|
0.100 |
12.36 |
2.8339 |
0.2834 |
35.0270 |
0.0283 |
3.5027 |
|
0.200 |
24.83 |
0.2313 |
0.0463 |
5.7432 |
0.0093 |
1.1486 |
|
0.300 |
35.91 |
0.0671 |
0.0201 |
2.4096 |
0.0060 |
0.7229 |
|
0.400 |
48.79 |
0.0234 |
0.0094 |
1.1417 |
0.0037 |
0.4567 |
|
0.500 |
60.42 |
0.0104 |
0.0052 |
0.6284 |
0.0026 |
0.3142 |
Adding the values in the last four columns gives
\[\sum_{i}w_ix_i= 0.3644 \hspace{20px} \sum_{i}w_iy_i = 44.9499\]
\[\sum_{i}w_ix_i^2 = 0.0499 \hspace{20px} \sum_{i}w_ix_iy_i = 6.1451\]
Substituting these values into the Equation \(\ref{5.26}\) and Equation \(\ref{5.27}\) gives the estimated slope and estimated y-intercept as
\[b_1 = \dfrac{(6×6.1451)-(0.3644×44.9499)}{(6×0.0499)-(0.3644)^2}= 122.985\]
\[b_0 = \dfrac{44.9499-(122.985×0.3644)}{6} = 0.0224\]
The calibration equation is
\[S_\ce{std} = 122.98 × C_\ce{std} + 0.02\]
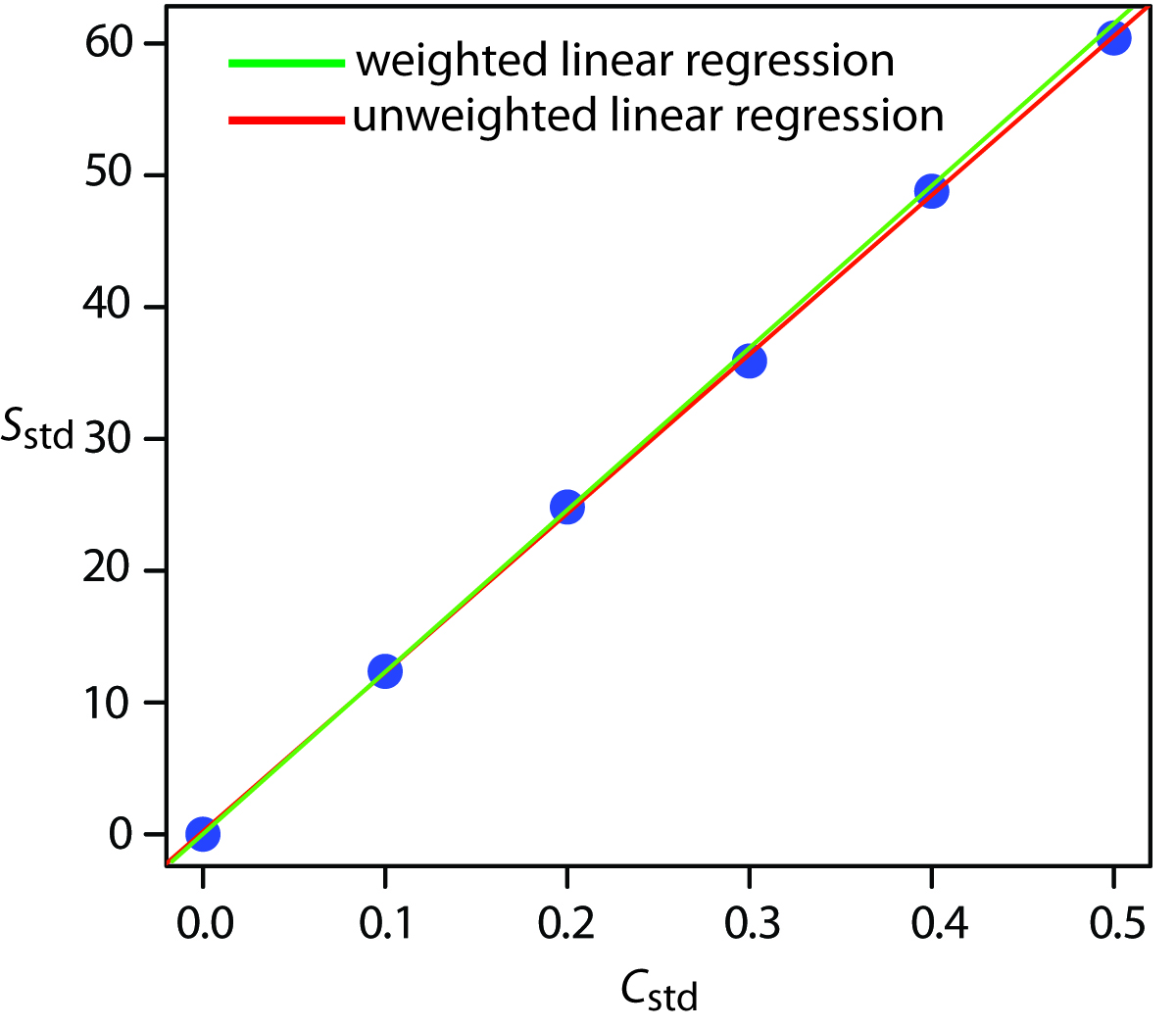
Figure 5.14 shows the calibration curve for the weighted regression and the calibration curve for the unweighted regression in Example 5.9. Although the two calibration curves are very similar, there are slight differences in the slope and in the y-intercept. Most notably, the y-intercept for the weighted linear regression is closer to the expected value of zero. Because the standard deviation for the signal, Sstd, is smaller for smaller concentrations of analyte, Cstd, a weighted linear regression gives more emphasis to these standards, allowing for a better estimate of the y-intercept.
Equations for calculating confidence intervals for the slope, the y-intercept, and the concentration of analyte when using a weighted linear regression are not as easy to define as for an unweighted linear regression.8 The confidence interval for the analyte’s concentration, however, is at its optimum value when the analyte’s signal is near the weighted centroid, yc, of the calibration curve.
\[y_c= \dfrac{1}{n \sum_i w_ix_i}\]
Figure 5.14: A comparison of unweighted and weighted normal calibration curves. See Example 5.9 for details of the unweighted linear regression and Example 5.12 for details of the weighted linear regression.
5.4.4 Weighted Linear Regression with Errors in Both x and y
If we remove our assumption that the indeterminate errors affecting a calibration curve exist only in the signal (y), then we also must factor into the regression model the indeterminate errors affecting the analyte’s concentration in the calibration standards (x). The solution for the resulting regression line is computationally more involved than that for either the unweighted or weighted regression lines.9 Although we will not consider the details in this textbook, you should be aware that neglecting the presence of indeterminate errors in x can bias the results of a linear regression.
See Figure 5.2 for an example of a calibration curve that deviates from a straight-line for higher concentrations of analyte.
5.4.5 Curvilinear and Multivariate Regression
A straight-line regression model, despite its apparent complexity, is the simplest functional relationship between two variables. What do we do if our calibration curve is curvilinear—that is, if it is a curved-line instead of a straight-line? One approach is to try transforming the data into a straight-line. Logarithms, exponentials, reciprocals, square roots, and trigonometric functions have been used in this way. A plot of log(y) versus x is a typical example. Such transformations are not without complications. Perhaps the most obvious complication is that data with a uniform variance in y will not maintain that uniform variance after the transformation.
Another approach to developing a linear regression model is to fit a polynomial equation to the data, such as y = a + bx + cx2. You can use linear regression to calculate the parameters a, b, and c, although the equations are different than those for the linear regression of a straight line.10 If you cannot fit your data using a single polynomial equation, it may be possible to fit separate polynomial equations to short segments of the calibration curve. The result is a single continuous calibration curve known as a spline function.
It is worth noting that in mathematics, the term “linear” does not mean a straight-line. A linear function may contain many additive terms, but each term can have one and only one adjustable parameter. The function
\[y = ax + bx^2\]
is linear, but the function
\[y = ax^b\]
is nonlinear. This is why you can use linear regression to fit a polynomial equation to your data. Sometimes it is possible to transform a nonlinear function. For example, taking the log of both sides of the nonlinear function shown above gives a linear function.
\[\log(y) = \log(a) + b\log(x)\]
The regression models in this chapter apply only to functions containing a single independent variable, such as a signal that depends upon the analyte’s concentration. In the presence of an interferent, however, the signal may depend on the concentrations of both the analyte and the interferent
\[S = k_\ce{A}C_\ce{A} +k_\ce{I}C_\ce{I} + S_\ce{reag}\]
where kI is the interferent’s sensitivity and CI is the interferent’s concentration. Multivariate calibration curves can be prepared using standards that contain known amounts of both the analyte and the interferent, and modeled using multivariate regression.11
Check out the Additional Resources at the end of the textbook for more information about linear regression with errors in both variables, curvilinear regression, and multivariate regression.