Solutions 11A
- Page ID
- 92329
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)S11.1
Write the Hamiltonian for the \(H_2\) and \(H_2^+\) molecules and explain each term. What terms differ between these two systems?
\[\hat{H}_{H_2^+}(1) = \dfrac{-\hbar^2}{2m_{e1}}\nabla_{e1}^2 + \dfrac{e^2}{4\pi \epsilon_0 R} - \dfrac{e^2}{4\pi \epsilon_0} \left(\frac{1}{r_{1A}} + \dfrac{1}{r_{1B}} \right) \]
In the \(H_2^+\) hamiltonian the first term is the kinetic energy term for the electron, the second term is the coulomb repulsion between the nuclei, and the last terms contain the electron nuclei attraction terms.
\[\hat{H}_{H_2}(1,2) = \dfrac{-\hbar^2}{2m_{e1}}\nabla_{e1}^2 + \dfrac{-\hbar^2}{2m_{e2}}\nabla_{e2}^2 + \dfrac{e^2}{4\pi \epsilon_0 R} - \dfrac{e^2}{4\pi \epsilon_0} \left(\dfrac{1}{r_{1A}} + \dfrac{1}{r_{1B}} + \dfrac{1}{r_{2A}} + \dfrac{1}{r_{2B}}\right) + \dfrac{e^2}{4\pi \epsilon_0 r_{12}}\]
In the \(H_2\) Hamiltonian the first two terms are the kinetic terms for each electron, the third term is coulomb repulsion between nuclei A and B which under the Born-Oppenhiemer approximation is static, and the last terms contain the electron nuclei attraction and the electron and electron repulsion term.
The difference between the two Hamiltonians is that with the loss of the second electron you lose the kinetic and potential terms related to that electron as well as the electron-electron repulsion term.
S11.2
Use normalization to identify the constants (\(c_1,c_2)\) for the LCAO approximation for the two molecular orbitals discussed in class \[\psi^+ = c_1\phi_A + c_2\phi_B\] and \[\psi^- = c_1\phi_A - c_2\phi_B\] with \(\phi\) is the 1s atomic orbital on the \(A\) and \(B\) nuclei, respectively.
The first thing to notice is that |c1|2 = |c2|2 since the electron should have no preference on which proton it is near, so their probabilities should be the same.
\[\langle \psi_{\pm} | \psi_{\pm} \rangle = |c_{\pm}|^2 \left(\int \phi_A^2 d\tau + \int \phi_B^2 d\tau \pm \int \phi_B \phi_A d\tau \pm \int \phi_A \phi_B d\tau \right) = 1\]
and
\[\int \phi_A^2 d\tau = \int \phi_B^2 d\tau = 1\]
since they are normalized atomic wavefunctions.
\[\langle \psi_{\pm} | \psi_{\pm} \rangle = |c_{\pm}|^2 (1 + 1 \pm (S + S^*)) = 1\]
Since both wavefunctions are real \(S = S^*\)
\[\langle \psi_{\pm} | \psi_{\pm} \rangle = |c_{\pm}|^2 (1 + 1 \pm 2S) = 1\]
\[|c_{\pm}| = (2 \pm 2S)^{-1/2}\]
S11.3
\(\psi^+\) and \(\psi^-\) are called the bonding and antibonding molecular orbitals. Which is higher in energy based on the nature of the electron density distribution? Which molecular orbital has a node and where?
Based on the electron density (Figure 9.3.1b) the ψ− orbital is higher in energy since it has more probability density on the outside of the nuclei while ψ+ has more density in the middle of the nuclei. When the electron is not in the middle of the nuclei there is no shielding between the two protons which repel each other creating a higher energy situation. ψ− is the orbital with a node in the middle of the two nuclei which make sense since it is the higher energy orbital.
S11.4
Using your favorite plotting software, plot the following:
- Plot the normalized wavefunction for the bonding and antibonding orbitals for the H2+ as a function of internuclear distance, R (Re=106 pm).
- Plot the overlap integral, S as a function of R/a0.
- Plot the Coulomb integral, J as a function of R/a0.
- Plot the Exchange integral, K as a function of R/a0.
- Plot the energies of the H2+ molecule as a function of internuclear distance, R.
What is the most stable separation between these two atoms?
These should look like the curves in Lecture 26 for the expressions for the integrals as a function of internuclear distance.
S11.5
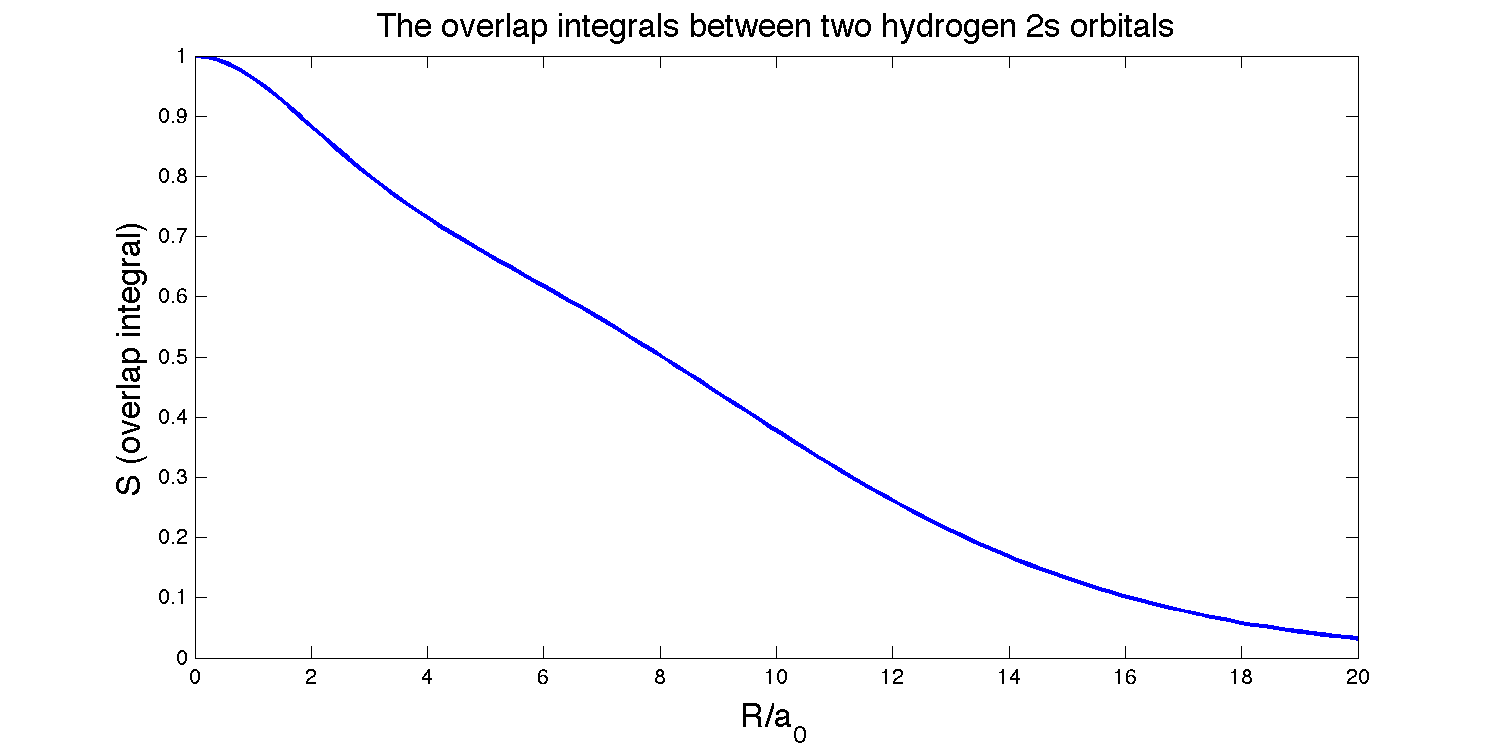
Derive the overlap integral between an \(H_{2s}\) orbital and a \(H_{2s}\) orbital on hydrogen nuclei separated by a distance \(R\). Plot this function and find the separation for which the overlap is a maximum.
The overlap integral between an \(H_{2s}\) orbital and a \(H_{2s}\) orbital on nuclei separated by a distance \(R\) is:
\[S(2s,2s)=\left\{1 + \dfrac{R}{2a_0} + \dfrac{1}{12}\left(\dfrac{R}{a_0}\right)^2 + \dfrac{1}{240}\left(\dfrac{R}{a_0}\right)^4\right\} e^{-R/2a}\]

Created by Ghunbong Cheung in Adobe Flash CS6
To find where the overlap is at its maximum, we set
\[\dfrac{dS}{dR} = 0\]
\[-\dfrac{Re^{-R/{2a_0}}(40a_0^3+20a_0^2R-8a_0R^2+R^3)}{480a_0^5} = 0\]
There are four roots for \(R\) since it is a quartic polynomial, but only one logical solution:
\[R = 0\]
Created by Ghunbong Cheung in MATLAB
This solution makes sense because these two orbitals are identical, so the maximum overlap should logical occur when the orbitals are exactly on top of each other.
S11.6
Contrast the bond orders for \(H_2^+\) and \(H_2\). The bond dissociation energy of \(H_2\) is 436 kJ/mol, which is less than twice that of \(H_2^+\); Why?
The bond order for \(H_2^+\) is \(1/2\) as there is a single bonding electron. For \(H_2\) the bond order is 1. So with double the bond order you might expect \(H_2\) to have double the dissociation energy. However \( 436 < 2D_{H_2^+} \) so in \(H_2\) there is an additional term in the Hamiltonian not present in \(H_2^+\) which is reducing the well depth and the dissociation energy. The nuclear repulsion is already accounted for in \(H_2^+\) but the electron repulsion is not. This additional repulsion factor is responsible for the change in dissociation energy between \(H_2^+\) and \(H_2\).
Another way to look at this that the one electron shields the nuclear charge of the other.
S11.7
If a electron-sensitive probe of volume 2.00 pm3 is inserted into an H2+ molecule-ion in its ground state, what is the probability of finding the electron:
- at nucleus A
- at nucleus B
- halfway between A and B
- at a point 40 pm along the bond from A and 15 pm perpendicularly off the bond
Do (a)-(d) again after the electron has been excited into the antibonding LCAO-MO.
Consider R = 106 pm.
Probability of finding an electron is given by \(P = \Big|\psi^2\Big|d\tau\).
We know that \(R = 106\;\text{pm}\) for H2+.
For the bonding LCAO-MO,
\[\psi_+^2=C_+^2(1s_A+1s_B)^2\]
\[\psi_+^2=C_+^2\dfrac{1}{\pi a_0^3}\Big(e^{-r_A/a_0}+e^{-r_B/a_0}\Big)^2\]
\[C_+^2\dfrac{1}{\pi a_0^3} = (0.561)^2\times\left(\dfrac{1}{\pi\times(52.9\;\text{pm})^3}\right)=6.7672\times10^{-7}\;\text{pm}^{-3}\]
(a) at nucleus A, \(r_A=0\) and \(r_B = R\):
\[\psi_+^2=8.72\times10^{-7}\;\text{pm}^{-3}\]
\[P = (8.72\times10^{-7}\;\text{pm}^{-3})\times(2.00\;\text{pm}^3) = 1.72\times10^{-6}\]
(b) at nucleus B:
\[P = 1.72\times10^{-6}\]
due to symmetry
(c) halfway between A and B, \(r_A=\dfrac{1}{2}R\) and \(r_B=\dfrac{1}{2}R\)
\[\psi_+^2=3.7\times10^{-7}\;\text{pm}^{-3}\]
\[P = \Big(3.7\times10^{-7}\;\text{pm}^{-3}\Big)\times(2.00\;\text{pm}^3) = 7.4\times10^{-7}\]
(d) at a point 40 pm along the bond from A and 15 pm perpendicularly, \(r_A=42.7\;\text{pm}\) and \(r_B=67.7\;\text{pm}\), these values are found using Pythagorean Theorem.
\[\psi_+^2=(e^{-42.7/52.9}+e^{-67.7/52.9})^2\times(6.7672\times10^{-7}\;\text{pm}^{-3})=3.55\times10^{-7}\;\text{pm}^{-3}\]
\[P = 3.55\times10^{-7}\;\text{pm}^{-3}\times(2.00\;\text{pm}^3)=7.09\times10^{-7}\]
For the antibonding LCAO-MO
\[\psi_-^2=C_-^2(1s_A-1s_B)^2\]
\[\psi_-^2=C_-^2\dfrac{1}{\pi a_0^3}\Big(e^{-r_A/a_0}-e^{-r_B/a_0}\Big)^2\]
\[C_-^2\dfrac{1}{\pi a_0^3} = (1.10)^2\times\Big(\dfrac{1}{\pi\times(52.9\;\text{pm})^3}\Big)=2.6018\times10^{-7}\;\text{pm}^{-3}\]
(a) at nucleus A, \(r_A=0\) and \(r_B = R\):
\[\psi_-^2=19.6\times10^{-7}\;\text{pm}^{-3}\]
\[P = (19.6\times10^{-7}\;\text{pm}^{-3})\times(2.00\;\text{pm}^3) = 3.92\times10^{-6}\]
(b) at nucleus B:
\[P = 3.92\times10^{-6}\]
due to symmetry
(c) halfway between A and B, \(r_A=\dfrac{1}{2}R\) and \(r_B=\dfrac{1}{2}R\)
\[\psi_-^2=0\]
\[P = 0\]
(d) at a point 40 pm along the bond from A and 15 pm perpendicularly, \(r_A=42.7\;\text{pm}\) and \(r_B=67.7\;\text{pm}\), these values are found using Pythagorean Theorem.
\[\psi_-^2=(e^{-42.7/52.9}-e^{-67.7/52.9})^2\times(2.6018\times10^{-7}\;\text{pm}^{-3})=7.20\times10^{-8}\;\text{pm}^{-3}\]
\[P = 7.20\times10^{-8}\;\text{pm}^{-3}\times(2.00\;\text{pm}^3)=1.44\times10^{-7}\]
S11.8
Calculate the bond orders and spin multiplicity of the following molecules:
- B2
- O2
- N2
- N2+
- H2+
- \( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^2 (\sigma^*_{2s})^2(\pi_{2p})^2 \)
- \( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^1 (\sigma^*_{2s})^1 \)
- \( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^2 (\sigma^*_{2s})^2 (\sigma_{2p_z})^2 (\pi_{2p})^4(\pi^*_{2p})^2 \)
Which are paramagnetic?
| Species | Bond Order | Spin Multiplicity | Paramagnetic |
|---|---|---|---|
| B2 | 1 | 1 | no |
| O2 | 2 | 3 | yes |
| N2 | 3 | 1 | no |
| N2+ | 2.5 | 2 | yes (weakly) |
| H2+ | 0.5 | 2 | yes (weakly) |
|
\( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^2 (\sigma^*_{2s})^2(\pi_{2p})^2 \) |
1 | 1 | no |
|
\( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^1 (\sigma^*_{2s})^1 \) |
0 | 3 (assuming Hund's rule applies for this excited state) | yes (if the molecule exists) |
|
\( (\sigma_{1s})^2 (\sigma^*_{1s})^2(\sigma_{2s})^2 (\sigma^*_{2s})^2 (\sigma_{2p_z})^2 (\pi_{2p})^4(\pi^*_{2p})^2 \) (This is isoelectronic with \(O_2\)) |
2 | 3 | yes |
S11.9
Which of the three species is the is the least stable due to bond order: O2, O2+, O22-. Hint it may be helpful to draw molecular orbitals for each species although it is not required.
We have to use the bond order formula for each of the situations.
\[\text{Bond order} = \dfrac{(\text{number of bonding electrons})- (\text{number of anti-bonding electrons})}{2} \]
- For O2: We have 12 valence electrons. Therefore
\[\text{Bond order} = \dfrac{10 - 6}{2} = 2\]
- For O2+: We have 11 valence electrons. Therefore
\[\text{Bond order} = \dfrac{10 - 5}{2} = \dfrac{5}{2}\]
- For O22-: We have 14 valence electrons. Therefore
\[\text{Bond order}= \dfrac{10 - 8}{2} = 1 \]
From the bond orders, we can see that O22- is the least stable. A similar problems is also outline here in Lecture 27: Molecular Oxygen is Paramagnetic. If you desire a similar exercises to test your skills, substitute \(N\) for \(O\) in the problem and see how things change.
S11.10
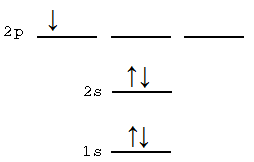
Draw an electronic energy level arrow diagram (i.e., the electronic configuration with levels and spins either up or down per electron) for this secular determinant:
\[\Psi(1,2,3,4,5) =\begin{vmatrix} 1s \alpha(1) & 1s \beta(1) & 2s \alpha(1) & 2s \beta(1) & 2p_x \beta(1) \\ 1s \alpha(2) & 1s \beta(2) & 2s \alpha(2) & 2s \beta(2) & 2p_x \beta(2) \\ 1s \alpha(3) & 1s \beta(3) & 2s \alpha(3) & 2s \beta(3) & 2p_x \beta(3) \\ 1s \alpha(4) & 1s \beta(4) & 2s \alpha(4) & 2s \beta(4) & 2p_x \beta(4) \\ 1s \alpha(5) & 1s \beta(5) & 2s \alpha(5) & 2s \beta(5) & 2p_x \beta(5) \end{vmatrix} \]
This is a five electron wavefunctions that may be assigned to an atom, not a molecule. This is because the orbital components of the determinant are orbitals are atomic, not molecular. For example,, this determinate wavefunction may be the \(B\) atom (although it could be an ion too) and corresponds to the following electron configuration: