13: The Flow of Genetic Information
- Page ID
- 165237
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In the preceding sections, we have discussed the replication of the cell's DNA and the mechanisms by which the integrity of the genetic information is carefully maintained. What do cells do with this information? How does the sequence in DNA control what happens in a cell? If DNA is a giant instruction book containing all of the cell's "knowledge" that is copied and passed down from generation to generation, what are the instructions for? And how do cells use these instructions to make what they need?
You have learned in introductory biology courses that genes, which are instructions for making proteins, are made of DNA. You also know that information in genes is copied into temporary instructions called messenger RNAs that direct the synthesis of specific proteins. This description of flow of information from DNA to RNA to protein, shown on the previous page, is often called the Central Dogma of molecular biology and is a good starting point for an examination of how cells use the information in DNA.
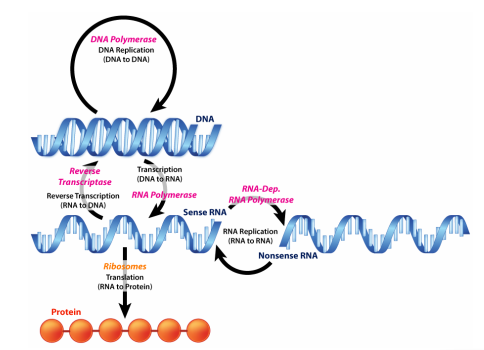 Figure 5.3.1: Central dogma
Figure 5.3.1: Central dogma
Consider that all of the cells in a multicellular organism have arisen by division from a single fertilized egg and therefore, all have the same DNA. Division of that original fertilized egg produces, in the case of humans, over a trillion cells, by the time a baby is produced from that egg (that's a lot of DNA replication!). Yet, we also know that a baby is not a giant ball of a trillion identical cells, but has the many different kinds of cells that make up tissues like skin and muscle and bone and nerves. How did cells that have identical DNA turn out so different.
The answer lies in gene expression, which is the process by which the information in DNA is used. Although all the cells in a baby have the same DNA, each different cell type uses a different subset of the genes in that DNA to direct the synthesis of a distinctive set of RNAs and proteins. The first step in gene expression is transcription, which we will examine next.
What is transcription? Transcription is the process of copying information from DNA sequences into RNA sequences. This process is also known as DNA-dependent RNA synthesis. When a sequence of DNA is transcribed, only one of the two DNA strands is copied into RNA.
 Figure 5.3.2: General Features of Transcription
Figure 5.3.2: General Features of Transcription
But, apart from copying one, rather than both strands of DNA, how is transcription different from replication of DNA. DNA replication serves to copy all the genetic material of the cell and occurs before a cell divides, so that a full copy of the cell's genetic information can be passed on to the daughter cell. Transcription, by contrast, copies short stretches of the coding regions of DNA to make RNA. Different genes may be copied into RNA at different times in the cell's lifecycle. RNAs are, so to speak, temporary copies of instructions of the information in DNA and different sets of instructions are copied for use at different times.
 Figure 5.3.3: RNA structure
Figure 5.3.3: RNA structure
Cells make several different kinds of RNA:
- mRNAs that code for proteins
- rRNAS that form part of ribosomes
- tRNAs that serve as adaptors between mRNA and amino acids during translation
- Micro RNAs that regulate gene expression
- Other small RNAs that have a variety of functions.
Building an RNA strand is very similar to building a DNA strand. This is not surprising, knowing that DNA and RNA are very similar molecules. What enzyme carries out transcription? Transcription is catalyzed by the enzyme RNA Polymerase. "RNA polymerase" is a general term for an enzyme that makes RNA. There are many different RNA polymerases.
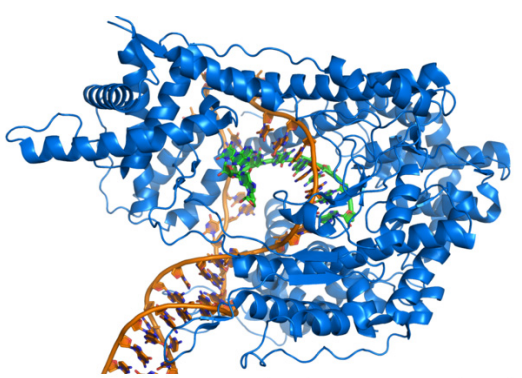 Figure 5.3.4: T7 RNA Polymerase (blue) making RNA (green) using DNA template (brown)
Figure 5.3.4: T7 RNA Polymerase (blue) making RNA (green) using DNA template (brown)
Like DNA polymerases, RNA polymerases synthesize new strands only in the 5' to 3' direction, but because they are making RNA, they use ribonucleotides (i.e., RNA nucleotides) rather than deoxyribonucleotides. Ribonucleotides are joined in exactly the same way as deoxyribonucleotides, which is to say that the 3'OH of the last nucleotide on the growing chain is joined to the 5' phosphate on the incoming nucleotide.
One important difference between DNA polymerases and RNA polymerases is that the latter do not require a primer to start making RNA. Once RNA polymerases are in the right place to start copying DNA, they just begin making RNA by stringing together RNA nucleotides complementary to the DNA template.
This, of course, brings us to an obvious question- how do RNA polymerases "know" where to start copying on the DNA. Unlike the situation in replication, where every nucleotide of the parental DNA must eventually be copied, transcription, as we have already noted, only copies selected genes into RNA at any given time.
Consider the challenge here: in a human cell, there are approximately 6 billion basepairs of DNA. Most of this is non- coding DNA, meaning that it won't need to be transcribed. The small percentage of the genome that is made up of coding sequences still amounts to between 20,000 and 30,000 genes in each cell. Of these genes, only a small number will need to be expressed at any given time.
What indicates to an RNA polymerase where to start copying DNA to make a transcript? Signals in DNA indicate to RNA polymerase where it should start (and end) transcription. These signals are special sequences in DNA that are recognized by the RNA polymerase or by proteins that help RNA polymerase determine where it should bind the DNA to start transcription. A DNA sequence at which the RNA polymerase binds to start transcription is called a promoter.
A promoter is generally situated upstream of the gene that it controls. What this means is that on the DNA strand that the gene is on, the promoter sequence is "before" the gene. Remember that, by convention, DNA sequences are read from 5' to 3'. So the promoter lies 5' to the start point of transcription.
Also notice that the promoter is said to "control" the gene it is associated with. This is because expression of the gene is dependent on the binding of RNA polymerase to the promoter sequence to begin transcription. If the RNA polymerase and its helper proteins do not bind the promoter, the gene cannot be transcribed and it will therefore, not be expressed.
 Figure 5.3.5: Promoter Sequences
Figure 5.3.5: Promoter Sequences
What is special about a promoter sequence? In an effort to answer this question, scientists looked at many genes and their surrounding sequences. It makes sense that because the same RNA polymerase has to bind to many different promoters, the promoters should have some similarities in their sequences. Sure enough, common sequence patterns were seen to be present in many promoters. We will first take a look at prokaryotic promoters. When prokaryotic genes were examined, the following features commonly emerged (Figure 5.3.5):
- A transcription start site (this the base in the DNA across from which the first RNA nucleotide is paired).
- A -10 sequence: this is a 6 bp region centered about 10 bp upstream of the start site. The consensus sequence at this position is TATAAT. In other words, if you count back from the transcription start site, which by convention, is called the +1, the sequence found at -10 in the majority of promoters studied is TATAAT).
- A -35 sequence: this is a sequence at about 35 basepairs upstream from the start of transcription. The consensus sequence at this position is TTGACA.
What is the significance of these sequences? It turns out that the sequences at -10 and -35 are recognized and bound by a subunit of prokaryotic RNA polymerase before transcription can begin.
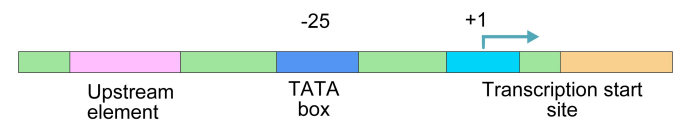 Figure 5.3.6: Promoter sequence elements
Figure 5.3.6: Promoter sequence elements
The RNA polymerase of E. coli, for example, has a subunit called the sigma subunit (or sigma factor) in addition to the core polymerase, which is the part of the enzyme that actually makes RNA. Together, the sigma subunit and core polymerase make up what is termed the RNA polymerase holoenzyme. The sigma subunit of the polymerase (shown in brown in Figure 5.3.7) can recognize and bind to the -10 and -35 sequences in the promoter, thus positioning the RNA polymerase (shown in green) at the right place to initiate transcription. Once transcription begins, the core polymerase and the sigma subunit separate, with the core polymerase continuing RNA synthesis and the sigma subunit wandering off to escort another core polymerase molecule to a promoter. The sigma subunit can be thought of as a sort of usher that leads the polymerase to its "seat" on the promoter.
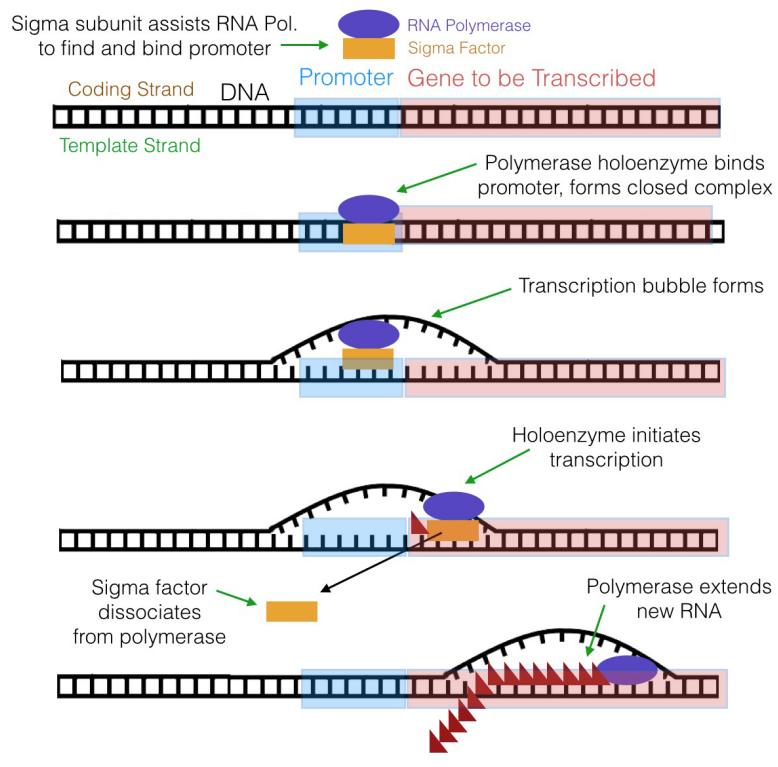 Figure 5.3.7: Transcription initiation in E. coli
Figure 5.3.7: Transcription initiation in E. coli
As already mentioned, an RNA chain, complementary to the DNA template, is built by the RNA polymerase by the joining of the 5' phosphate of an incoming ribonucleotide to the 3'OH on the last nucleotide of the growing RNA strand. How does the polymerase know where to stop? A sequence of nucleotides called the terminator is the signal to the RNA polymerase to stop transcription and dissociate from the template.
Although the process of RNA synthesis is the same in eukaryotes as in prokaryotes, there are some additional issues to keep in mind in eukaryotes. One is that in eukaryotes, the DNA template exists as chromatin, where the DNA is tightly associated with histones and other proteins. The "packaging" of the DNA must therefore be opened up to allow the RNA polymerase access to the template in the region to be transcribed.
A second difference is that eukaryotes have multiple RNA polymerases, not one as in bacterial cells. The different polymerases transcribe different genes. For example, RNA polymerase I transcribes the ribosomal RNA genes, while RNA polymerase III copies tRNA genes. The RNA polymerase we will focus on most is RNA polymerase II, which transcribes protein-coding genes to make mRNAs.
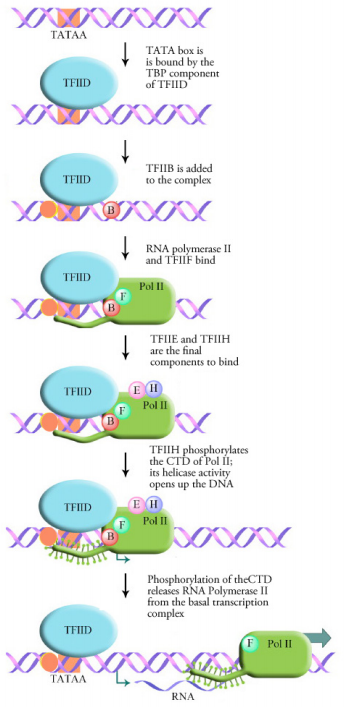 Figure 5.3.8: Assembly of basal transcription complex and initiation of transcription
Figure 5.3.8: Assembly of basal transcription complex and initiation of transcription
All three eukaryotic RNA polymerases need additional proteins to help them get transcription started. In prokaryotes, RNA polymerase by itself can initiate transcription (remember that the sigma subunit is a subunit of the prokaryotic RNA polymerase). The additional proteins needed by eukaryotic RNA polymerases are referred to as transcription factors. We will see below that there are various categories of transcription factors.
Finally, in eukaryotic cells, transcription is separated in space and time from translation. Transcription happens in the nucleus, and the mRNAs produced are processed further before they are sent into the cytoplasm. Protein synthesis (translation) happens in the cytoplasm. In prokaryotic cells, mRNAs can be translated as they are coming off the DNA template, and because there is no nucleus, transcription and protein synthesis occur in a single cellular compartment.
Like genes in prokaryotes, eukaryotic genes also have promoters. Eukaryotic promoters commonly have a TATA box, a sequence about 25 basepairs upstream of the start of transcription that is recognized and bound by proteins that help the RNA polymerase to position itself correctly to begin transcription. (Some eukaryotic promoters lack TATA boxes, and have, instead, other recognition sequences to help the RNA polymerase find the spot on the DNA where it spot on the DNA where it binds and initiates transcription.)
We noted earlier that eukaryotic RNA polymerases need additional proteins to bind promoters and start transcription. What are these additional proteins that are needed to start transcription? General transcription factors are proteins that help eukaryotic RNA polymerases find transcription start sites and initiate RNA synthesis. We will focus on the transcription factors that assist RNA polymerase II. These transcription factors are named TFIIA, TFIIB and so on (TF= transcription factor, II=RNA polymerase II, and the letters distinguish individual transcription factors).
Transcription in eukaryotes requires the general transcription factors and the RNA polymerase to form a complex at the TATA box called the basal transcription complex or transcription initiation complex. This is the minimum requirement for any gene to be transcribed. The first step in the formation of this complex is the binding of the TATA box by a transcription factor called the TATA Binding Protein or TBP. Binding of the TBP causes the DNA to bend at this spot and take on a structure that is suitable for the binding of additional transcription factors and RNA polymerase. As shown in the figure at left, a number of different general transcription factors, together with RNA polymerase (Pol II) form a complex at the TATA box.
The final step in the assembly of the basal transcription complex is the binding of a general transcription factor called TFIIH. TFIIH is a multifunctional protein that has helicase activity (i.e., it is capable of opening up a DNA double helix) as well as kinase activity. The kinase activity of TFIIH adds a phosphate onto the C-terminal domain (CTD) of the RNA polymerase. This phosphorylation appears to be the signal that releases the RNA polymerase from the basal transcription complex and allows it to move forward and begin transcription.


