4: Understanding Public Chemical Databases
- Page ID
- 95657
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\) |
CHEM3351: Cheminformatics Spring 2018: Bucholtz LibreText |
 |
Learning Objectives
- Explain what primary and secondary databases are.
- Explain what data provenance is.
- Review publicly available chemical databases in different domains.
- Understand how PubChem data are organized.
- Learn how to critically assess data in public databases.
1. (SOME) DATABASE BASIC
1.1. What is a database?
A database is an “organized collection of information.” The information in a database can be in any format, including texts, numbers, images, audios, videos, and many others (and combination of these), but this information must be “organized” for efficient retrieval. According to this definition, a database is not necessarily electronic (i.e., accessible by computers). For example, the collection of names in a phone book or address book may also be considered as a database, because the names are arranged (typically in alphabetical order) to make it easy to search for necessary information (e.g., phone numbers or addresses). However, in computer science and related areas, a database usually means an electronic database. Therefore, the term “database” in this module is used to mean an “electronic database”.
1.2. Primary databases vs. secondary databases
Databases are often categorized into primary and secondary databases.
- Primary databases contain experimentally-derived data that are directly submitted by researchers (also called “primary data”). In essence, these databases serve as archives that keep original data. Therefore, they are also known as archival databases.
- Secondary databases contain secondary data, which are derived from analyzing and interpreting primary data. These databases often provide value-added information related to the primary data, by using information from other databases and scientific literature. Essentially, secondary databases serve as reference libraries for the scientific community, providing highly curated reviews about primary data. For this reason, they are also known as curated databases, or knowledgebase.
It should be noted that the distinction between primary and secondary databases is not always clear and that many databases have the characteristics of both primary and secondary database. It is very common that a primary database curates its data with information drawn from secondary databases. In addition, because many secondary databases make their value-added information available in the public domain, data exchange and integration among databases very frequently occurs. As a results, virtually all data providers also becomes data consumers these days.
1.3. Data provenance
The term “data provenance” refers to a record trail that describes the origin or source of a piece of data and the process by which it entered in a database.1 Simply put, data provenance deals with the questions “where the data came from” and “how and why the data is in its present place”. Although the data provenance information is critical in the reliability of a data source (and its data), this information is not easy to manage. In addition, information predicted in one database may not be appropriate for use in other databases, but may end up being integrated in them anyway. Therefore, databases need to document the provenance of the data and devise a way to notify users of that information. In turn, users should always pay attention to the data provenance issue when using a database.
2. PUBLIC CHEMICAL DATABASE
These days many public online databases provide chemical information free of charge and the databases mentioned in this module are only a few examples of them. Note that these databases vary in size and scope.
2.1. PubChem: chemical information repository at the U.S. NIH
PubChem (https://pubchem.ncbi.nlm.nih.gov)2-4 is a public repository of information on small molecules and their biological activities, developed and maintained by the National Library of Medicine (NLM), an institute within the U.S. National Institutes of Health (NIH). Since its launch in 2004 as a component of the NIH’s Molecular Libraries Roadmap Initiatives, it has been rapidly growing, and now serves as a key chemical information resource for researchers in many biomedical science areas, including cheminformatics, chemical biology, and medicinal chemistry. Detailed information on PubChem can be found in these three papers:
- PubChem Substance and Compound databases
S. Kim et al., Nucleic Acids Research 2016, 44, D1202-D1213
(https://doi.org/10.1093/nar/gkv951)
- PubChem BioAssay: 2017 update
Wang Y. et al. Nucleic Acids Research 2017, 45, D955-D963
(https://doi.org/10.1093/nar/gkw1118) - Getting the most out of PubChem for virtual screening
S. Kim, Expert Opin. Drug Discov. 2016, 11, 843-855
(http://dx.doi.org/10.1080/17460441.2016.1216967) (Available to logged-in students at bottom of this module.)
As of February 2017, PubChem contains more than 235 million depositor-provided substances, 94 million unique chemical structures, and one million biological assays, which cover about 10 thousand protein target sequences. For efficient use of this vast amount of data, PubChem provides various search and analysis tools. Some of these search tools will be used later in this course for demonstration purposes.
2.2. ChemSpider: a chemical database integrated with RSC’s publishing process
ChemSpider (http://www.chemspider.com/)5,6 is a free chemical structure database, containing information on 34 million structures collected from ~500 data sources. It also provides information on chemical reactions through ChemSpider SyntheticPages (CSSP)7. ChemSpider uses a crowdsourcing approach that allows registered users for manual comment and correction of ChemSpider records. Owned by the Royal Society of Chemistry (RSC), which publishes ~40 peer-reviewed chemistry journals, ChemSpider is integrated with the RSC publishing process, whereby new chemicals identified in newly published RSC articles also become available in ChemSpider.
2.3. ChEMBL: literature-extracted biological activity information
ChEMBL (https://www.ebi.ac.uk/chembl/)8,9 is a large bioactivity database, developed and maintained by the European Bioinformatics Institute (EBI), which is part of the European Molecular Biology Laboratory (EMBL). The core activity data in ChEMBL are “manually” extracted from the full text of peer-reviewed scientific publications in select chemistry journals, such as Journal of Medicinal Chemistry, Bioorganic Medicinal Chemistry Letters, and Journal of Natural products. From each publication, details of the compounds tested, the assays performed and any target information for these assays are abstracted. ChEMBL also integrates screening results and bioactivity data from other public databases (such as PubChem BioAssay) and information on approved drugs from the U.S. FDA Orange Book10and the NLM’s DailyMed11.
2.4. ChEBI: a dictionary of small molecular entity
ChEBI (https://www.ebi.ac.uk/chebi/)12,13 stands for “Chemical Entities of Biological Interest”. It is a freely available database of “small” molecular entities, developed at the European Bioinformatics Institute (EBI). The molecular entities in ChEBI are either natural or synthetic products used to intervene the processes of living organisms. As a rule, however, ChEBI does not contain macromolecules directly encoded by genome (e.g., nucleic acids, proteins, and peptides derived from protein by cleavage). ChEBI provides “standardized” descriptions of molecular entities that enable other databases to annotate their entries in a consistent fashion. ChEBI focuses on high-quality manual annotation, non-redundancy, and provision of a chemical ontology rather than full coverage of the vast chemical space. Note that both ChEMBL and ChEBI are developed and maintained by the EMBL-EBI. While ChEMBL focuses on “bioactivity” of a large number of bioactive molecules (currently ~2.0 millions), ChEBI is a “dictionary” that provides high-quality standardized descriptions for a relatively small number of molecules (currently ~50 thousands).
2.5. NIST Webbook: thermodynamic and spectroscopic data of chemicals
The U.S. National Institutes of Standards and Technology (NIST) compiles chemical and physical property data for chemical species and distributes them through the web site called the NIST Chemistry WebBook (http://webbook.nist.gov)14,15. These data include thermochemical data (e.g., enthalpy of formation, heat capacity, and vapor pressure), reaction thermochemistry data (e.g., enthalpy of reaction and free energy of reaction), spectroscopic data (e.g., IR and UV/Vis spectra), gas chromatographic data, ion energetics data, and so on.
2.6. DrugBank: comprehensive information on drug molecules
DrugBank16-18 (http://www.drugbank.ca/) is a comprehensive online database containing biochemical and pharmacological information about ~8,000 drug molecules, including U.S. Food and Drug Administration (FDA)-approved small-molecule drugs and biotech drugs (e.g., protein/peptide drugs) as well as experimental drugs. DrugBank provides a wide range of drug information, including drug targets, mechanism of action, adverse drug reactions, food-drug and drug-drug interactions, experimental and theoretical ADMET properties (i.e., Absorption, Distribution, Metabolism, Excretion, and Toxicity), and many others. Most of these data are curated from primary literature sources, by domain-specific experts and skilled biocurators.
2.7. HMDB: the Human Metabolome Database
The Human Metabolome Database (HMDB) (http://www.hmdb.ca)19-21 is comprehensive information on human metabolites and human metabolism data. This database contains curated information derived from scientific literature, as well as experimentally determined metabolite concentrations in human tissue or biofluid (e.g., urine, blood, cerebrospinal fluid and so on). Reference Mass spectra (MS) and nuclear magnetic resonance (NMR) spectra for metabolites are also provided when available. In addition to data for “detected” metabolites (those with measured concentrations or experimental confirmation of their existence), the HMDB also provides information on “expected” metabolites (those for which biochemical pathways are known or human intake/exposure is frequent but the compound has yet to be detected in the body).
2.8. TOXNET: a collection of toxicological information
TOXNET (http://toxnet.nlm.nih.gov/)22-25, maintained by the National Library of Medicine (NLM) at NIH, is a group of databases covering toxicology, hazardous chemicals, toxic releases, environmental and occupational health, risk assessment. Currently, 16 databases are integrated into the TOXNET system, and users can search all these databases either at once or individually. While all the 16 databases provide valuable information, three of them may be worth mentioning in the context of this course.
- ChemIDPlus26,27 is a dictionary of over 400,000 chemical records (names, synonyms, and structures) and provides access to the structure and nomenclature files used for the identification of chemical substances in the TOXNET system and other NLM databases.
- The Hazardous Substances Data Bank (HSDB)28,29 focuses on the toxicology of potentially hazardous chemicals, providing information on human exposure, industrial hygiene, emergency handling procedures, environmental fate, regulatory requirements, nanomaterials, and related areas. All HSDB data are referenced and derived from a core set of books, government documents, technical reports and selected primary journal literature. Importantly, HSDB is peer-reviewed by the Scientific Review Panel (SRP), a committee of experts in the major subject areas within the data bank's scope.
- The Comparative Toxicogenomics Database (CTD)30,31 contains manually curated data describing interactions of chemicals with genes/proteins and diseases. This database provides insight into the molecular mechanisms underlying variable susceptibility for environmentally influenced diseases.
A brief overview of TOXNET and its databases can be found in the TOXNET Fact Sheet23 and a recent paper by Fowler and Schnall25.
2.9. Protein Data Bank (PDB): a key source for protein-bound ligand structures
The Protein Data Bank (PDB) is an archive of the experimentally determined 3-D structures of large biological molecules such as proteins and nucleic acids. These structures were determined primarily by using X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy. While PDB is not a small molecule database, it contains the 3-D structures of many proteins with small-molecule ligands bound to them. PDB allows users to search for proteins that an input small molecule binds to. Considering that it is not possible to experimentally determine how small molecules (such as drug or toxic chemicals) actually bind to their target proteins in a living organism, PDB is the most widely used resource for experimentally determined protein-bound structures of small molecules. The PDB are maintained by the Worldwide PDB (wwPDB)32, and freely accessible via the websites of its member organizations: PDBe (PDB in Europe)33,34, PDBj (PDB Japan)35,36, RCSB PDB (Research Collaboratory for Structural Bioinformatics PDB)37,38.
3. DATA ORGANIZATION IN PUBCHEM AS A DATA AGGREGATOR
PubChem (https://pubchem.ncbi.nlm.nih.gov) is a data aggregator, meaning that it collects data from other data sources. As of February 2017, PubChem’s data are from more than 500 organizations, including government agencies, university labs, pharmaceutical companies, substance vendors, and other databases. An up-to-date list of PubChem’s data sources is available at the PubChem Sources page (https://pubchem.ncbi.nlm.nih.gov/sources). To better understand the features of this page, read this article on PubChem Blog:
PubChem organizes its data into three inter-linked databases: Substance, Compound, and BioAssay (See Table 1), which can be searched from either the PubChem home page (https://pubchem.ncbi.nlm.nih.gov) or the web page of one of the three PubChem databases.
|
Database |
URL |
Identifier |
|---|---|---|
|
Substance |
SID |
|
|
Compound |
CID |
|
|
BioAssay |
AID |
Individual data contributors deposit information on chemical substances to the Substance database (https://www.ncbi.nlm.nih.gov/pcsubstance). Different data contributors may provide information on the same molecule, hence the same chemical structure may appear multiple times in the Substance database. To provide a non-redundant view, chemical structures in the Substance database are normalized through a process called “standardization” and the unique chemical structures are identified and stored in the Compound database (https://www.ncbi.nlm.nih.gov/pccompound). The difference between the Substance and Compound databases is explained in more detail in this blog post.
Descriptions of biological experiments on chemical substances are stored in the BioAssay database (https://www.ncbi.nlm.nih.gov/pcassay). The unique identifiers used to locate records in these three databases are called SID (Substance ID), CID (Compound ID), and AID (Assay ID) for the Substance, Compound, and BioAssay databases, respectively.
All information in the Substance database is submitted by individual data depositors. However, the Compound database does contain information that are not submitted by data depositors, but annotated by the PubChem team. [In the context of scientific databases, annotation refers to the process of adding extra information to a database entry (for example, a compound in the Compound database and an assay in the BioAssay database)]. The annotated information is always presented with its provenance information (that is, the source of the information). The list of all the annotation sources used in PubChem is available at the PubChem Sources page (https://pubchem.ncbi.nlm.nih.gov/sources). From this page, one may download all the annotations from a particular source.
4. SPECIAL NOTES ON USING PUBLIC CHEMICAL DATABASES
All the databases mentioned above (including PubChem) are public databases that provide their contents free of charge, and in many cases they also provide a way to download data in bulk and integrate them into one’s own database. Therefore, it is very common that database groups exchange their information with each other. This often raises some technical concerns. For example, as mentioned in Part 3 of Module 2, different databases may use different chemical representations to refer to the same molecule. This may result in incorrect chemical structure matching between the databases, leading to incorrect data integration. In addition, when one database has incorrect information, this error often propagates into other databases. The error propagation issue is a serious, but very common, problem.39,40 Therefore, when using information in these databases, one should keep in mind various data accuracy and quality issues prevalent in these databases. A goal of this course is to help students develop the ability to critically assess chemical information available in public databases.
Questions
PART I. Getting familiar with PubChem
1. Read these articles and answer the following questions.
- What is the difference between a substance and a compound in PubChem?
(https://pubchemblog.ncbi.nlm.nih.gov/2014/06/19/what-is-the-difference-between-a-substance-and-a-compound-in-pubchem/) - Compound Summary Page Redesigned
https://pubchemblog.ncbi.nlm.nih.gov/2014/10/20/compound-summary-page-redesigned/) - Substance Record Page Released
(https://pubchemblog.ncbi.nlm.nih.gov/2015/04/09/substance-record-page-released/) - PubChem adds a “legacy” designation for outdated data
(https://pubchemblog.ncbi.nlm.nih.gov/2015/11/16/pubchem-adds-a-legacy-designation-for-outdated-data/) - “§2.4. Availability of compounds for subsequent experiments” in “Getting the most out of PubChem for virtual screening”
(http://www.tandfonline.com/doi/full/10.1080/17460441.2016.1216967) [If you don’t have access to this article, Author’s original manuscript for this paper is available as an attachment at the end of Module 4.]
- Explain the difference between the PubChem Substance and Compound databases in two or three sentences.
- Explain what the Compound Summary page of a compound is.
- Explain what the Substance Record page of a substance is.
- Explain the reason why the “legacy” designation was introduced in PubChem in two or three sentences.
- Among the menus available on the top of the PubChem home page (https://pubchem.ncbi.nlm.nih.gov) is “Today’s Statistics”. The number of compounds/substances/assays shown under this menu does not include “non-live” records. What does “non-live” mean here?
2. While the PubChem Substance database is an archive in nature, data providers often want to update their substance information archived in PubChem. For this reason, PubChem keeps all different “versions” of a substance record and shows the most recent version on its Substance Record page by default (Click here to read about what the Substance Record page is). Go to the PubChem home page (https://pubchem.ncbi.nlm.nih.gov) and follow the steps described below.
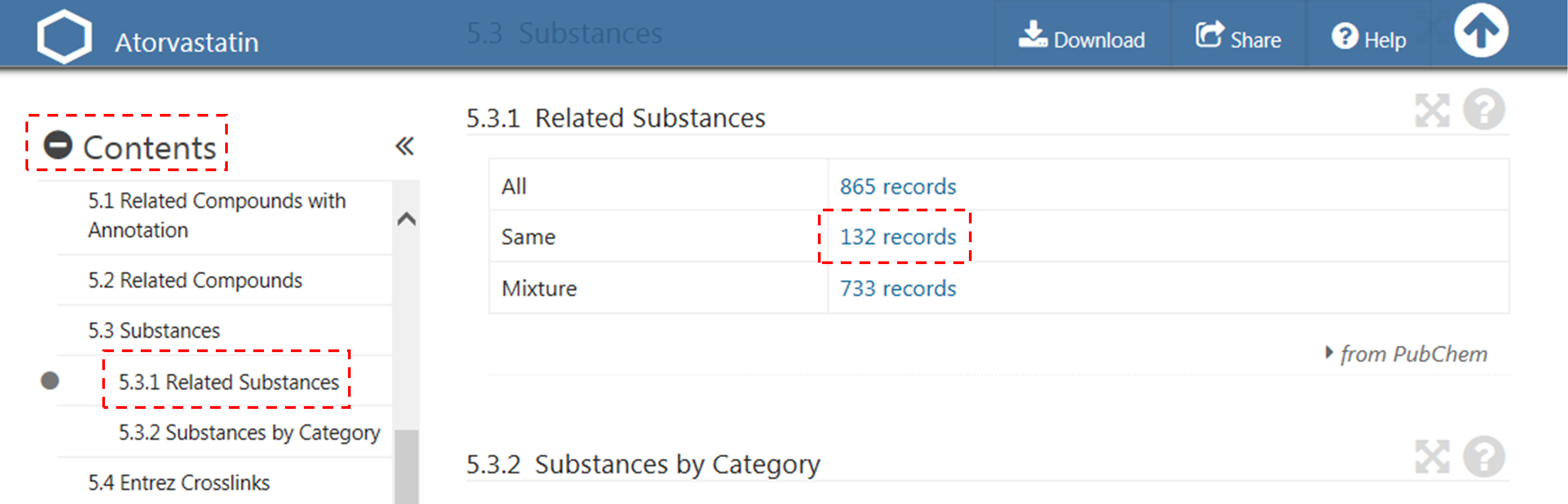
- After selecting the “Compound” tab above the search box, type “60823” in the search box and click the “Go” button. This will direct you to the Compound Summary page for CID 60823 (atorvastatin). (Click here to read about what the Compound Summary page is.) [You will learn how to search PubChem in much more detail for next two modules (Modules 5 and 6).]
- (Scroll down until you see “Contents” on the left column. Expand this table of contents by clicking the “+” sign before “Contents”. Locate the “Related Substances” section and click the record count for the “Same” item under that section.
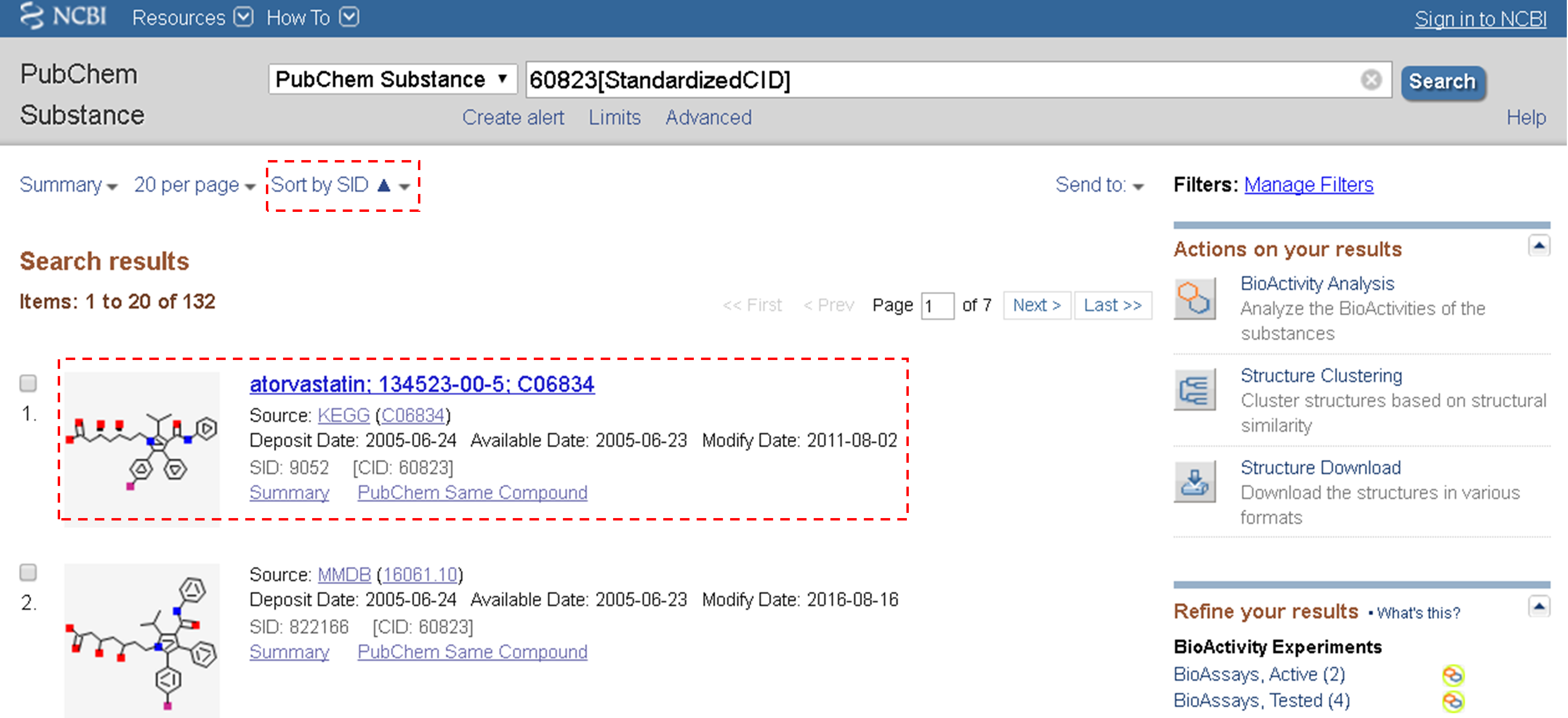
(c) The previous step will lead you to the web page that presents 132 substance records associated with CID 60823 (atorvastatin). This page is called a Document Summary (DocSum) page, because it presents a (very brief) summary of retrieved records. Sort the list by SID (in ascending order) and click the substance that appear on the top of the DocSum page (that is, SID 9052).
(d) Clicking SID 9052 directs you to the Substance Record page for SID 9052. From the Table of Contents, locate the “Modify Date” section. Now you see a table that shows a list of the dates when this substance record was modified.
(e) Click each version of this record and fill in the table below with the information under the “Depositor Comments” and “Cross-References” sections.
|
Version 1 |
Comments |
None |
|
RegID |
C06834 |
|
|
RN |
134523-00-5 |
|
|
DBURL |
||
|
SBURL |
||
|
Version 2 |
Comments |
|
|
RegID |
|
|
|
RN |
|
|
|
DBURL |
|
|
|
SBURL |
|
|
|
Version 3 |
Comments |
|
|
RegID |
|
|
|
RN |
|
|
|
DBURL |
|
|
|
SBURL |
|
|
|
Version 4 |
Comments |
|
|
RegID |
|
|
|
RN |
|
|
|
DBURL |
|
|
|
SBURL |
|
|
|
Version 5 |
Comments |
|
|
RegID |
|
|
|
RN |
|
|
|
DBURL |
|
|
|
SBURL |
|
|
|
Version 6 |
Comments |
Same as: D07474 |
|
RegID |
None |
|
|
RN |
None |
|
|
DBURL |
None |
|
|
SBURL |
None |
- The “Depositor Comments” section of version 6 of this record has a link to another substance record in PubChem. What is the SID of this substance record? What is the CID of the compound associated with this substance record? Is this CID the same as the one used in (a)?
- Briefly summarize (in three or four sentences) how depositor-provided information on SID 9052 has evolved over time.
3. Some records in PubChem are “non-live”, meaning that they are “not searchable”, although they do exist in the database. This exercise is designed to help students better understand what non-live records are.
- In the previous exercise, you searched the PubChem Compound database for CID “60823”. Now search the Compound database using the query “73336290”. What message do you get?
- Repeat the search using the query “60823” (which we already know returns a hit) to go to the Compound Summary page for CID 60823. What is the URL of this page (i.e., the web address)?
- Replace the string “60823” in the URL with “73336290” and press the “Enter” key on the keyboard. This will get you to the Compound Summary page of CID 73336290, which you were not able to find in (a). Indeed, this page presents the message “NOTE: NON-LIVE RECORD. See the related substances for more information.” How many substances are associated with CID 73336290? What are their SIDs?
- Go to the Substance Record page for the substance associated with CID 73336290 (by clicking the SID listed under the “Related Substances” section of its Compound Summary page). What is the version number of this substance record?
- Go to the Substance Record page for the most recent version of the substance record in (d). This page shows the message: “NOTE: REVOKED RECORD. See the revoke reason and the revision history for more information.” What is listed as the revoke reason for this record?
PART II. Finding information on caffeine
4. Go to the PubChem home page (https://pubchem.ncbi.nlm.nih.gov). After selecting the “Compound” tab above the search box, type “caffeine[CompleteSynonym]” in the search box and click the “Go” button. It will lead you to the Compound Summary page for caffeine (CID 2519), which presents all information available in PubChem for this chemical. [The suffix “[CompleteSynonym]” means that you want to search for compounds whose name exactly matches the query string (caffeine). You will learn how to search PubChem in much more detail for next two modules (Modules 5 and 6).]
(a) Scroll down until you see “Contents” on the left column. Expand this table of contents by clicking the “+” sign before “Contents”. Locate the “boiling point” and “melting point” sections (under “Experimental Properties” of “Chemical and Physical Properties”). Fill the table below using the information presented in these sections. Because the data in these sections are given in either ºC or ºF, you need to convert from one unit to the other to fill the table.
|
Primary Data source |
Boiling points in ºC |
Boiling points in ºF |
|---|---|---|
|
HSDB |
|
|
|
ILO-ICSC |
|
|
|
CAMEO Chemicals |
177.8°C |
352° F at 760 mm Hg (sublimes) (NTP, 1992) |
|
Data source |
Melting points in ºC |
Melting points in ºF |
|---|---|---|
|
HSDB |
|
|
|
DrugBank (Phys Prop) |
|
|
|
ILO-ICSC |
|
|
|
CAMEO Chemicals |
237.8 °C |
460° F (NTP, 1992) |
|
Human Metabolome Database |
|
|
- The reported boiling points of caffeine are actually the sublimation temperature, as indicated in parentheses. Explain the difference between boiling and sublimation.
- Suggest a reason why PubChem reports the sublimation temperature under the boiling point section?
- The sublimation temperature and melting point of caffeine summarized in the table does not seem reasonable if you compare them with each other. Explain why they are not reasonable.
- Below is the schematic phase diagram of caffeine. Based on this diagram, what kind of information do you need to resolve the data accuracy issue identified in (d).
- Among the data sources in the table, how many sources explicitly reported the atmospheric pressure at which the phase transition temperatures were measured? What are they?
- Whenever possible, PubChem presents data with its provenance (source). For each boiling and melting point value, click the source name to find more detailed information about the data provenance. In most case, PubChem provides a link to the data reported on the original data source. Click this link to check the melting and boiling point data on the data sources’ websites. Does any data source provide additional information about the boiling and melting points of caffeine that may help you figure out the issue with the phase transition temperatures of caffeine?
- Provide a short paragraph (less than 100 words) that evaluates the accuracy of the phase transition temperatures in PubChem and other databases, based on your answers to questions (a) ~ (g).
- The term “curation” refers to the process of critically reviewing the data as well as checking its accuracy in order to provide the data and related information for the scientific community. What you did while answering the questions (a)~(h) may be considered as a part of manual curation efforts, and the paragraph written in (h) is essentially a curated review of the phase transition temperature data of caffeine. Provide the total (estimated) time you spent for answering questions (a) ~ (h), and use it to estimate the amount of time necessary to manually curate the melting and boiling data for 10, 100, 1000, 10000 compounds, respectively.
- Curation can be done either manually or in an automated way. Discuss (in less than 50 words) the pros and cons of manual and automated curations and when each approach would be appropriate.
PART III. Understanding toxicity of caffeine
5. Go back to the PubChem Compound Summary page for caffeine (CID 2519). Locate the “GHS classification” section (under “Hazard Identification” of “Safety & Hazards”). This section shows the GHS (Globally Harmonized System of Classification and Labelling of Chemicals) information of caffeine collected from authoritative organizations. (Click here to read more about the GHS information.)
(a) By default, the “GHS classification” section shows the GHS information from a single organization. To view information from all organizations, click the “View GHS classification from all sources” at the end of the section. Based on the information on this page, fill in the table below.
|
EU Regulation & Austrailia |
Japan NITE |
ECHA |
|
|
Pictograms |
|
||
|
GHS hazard statement code |
H302 |
(b) What does each pictogram in the table mean? Refer to the PubChem GHS classification page (https://pubchem.ncbi.nlm.nih.gov/ghs/#_pict).
(c) What does each hazard statement code means? Refer to the PubChem GHS classification page (https://pubchem.ncbi.nlm.nih.gov/ghs/#_prec)
(d) According to the information collected in the table, caffeine has acute oral toxicity. However, from our experience, we know that the consumption of caffeine in a cup of coffee is usually okay. To better understand the GHS information on caffeine, it is necessary to review the oral toxicity data of caffeine. First fill in this table with the oral LD50 values for different animal species compiled in the “Non-Human Toxicity values” sections (under “Toxicological Information” of “Toxicity”).
|
Oral LD50 value |
|
|
Rat |
192 mg/kg |
|
Mouse |
|
|
Dog |
|
|
Rabbit |
|
|
Guinea pig |
|
|
Hamster |
(e) Fill in the table with the caffeine content in typical food and drinks, which can be found in the “Food Survey Values” section (under “Ecological Information” of “Toxicity”). To compare these values with the oral LD50 values, divide the caffeine content by the average bodyweight of adult humans (assumed to 60 kg). Also calculate the per cent value of the caffeine content per kg. relative to the oral LD50 value for mice [that is, caffeine content per kg divided by LD50 (mice)].
|
Caffeine content |
Caffeine content per kg |
Ratio |
|
|
A single cup of espresso (30 mL) |
64 mg |
1.1 mg/kg |
0.84 % |
|
A 8-oz cup of automatic drip coffee |
|
|
|
|
A cup of tea (camellia sinensis) |
|
|
|
|
Soft drinks per 12 oz |
|
|
|
|
Energy drinks per 12 oz |
|
|
|
(f) Find information on human toxicity of caffeine from the “Human Toxicity Values” section (under “Toxicological Information” of “Toxicity”). Compare this information with the data collected in (d) and (e).
(g) Read the following articles about caffeine.
- http://www.fda.gov/downloads/UCM200805.pdf
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3839443/pdf/hpj4801-5.pdf
- http://www.fda.gov/food/recallsoutbreaksemergencies/safetyalertsadvisories/ucm405787.htm
(h) Based on the information collected from (a) through (g), write a short paragraph (less than 100 words) about the toxicity of caffeine and caffeine-containing products.
References
- (1) Ram, S.; Liu, J. In SWPM'09 Proceedings of the First International Conference on Semantic Web in Provenance Management Washington, D.C. , 2009; Vol. 526, p 35.
- (2) Kim, S.; Thiessen, P. A.; Bolton, E. E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L. Y.; He, J. E.; He, S. Q.; Shoemaker, B. A.; Wang, J. Y.; Yu, B.; Zhang, J.; Bryant, S. H. Nucleic Acids Res. 2016, 44, D1202.
- (3) Wang, Y.; Bryant, S. H.; Cheng, T.; Wang, J.; Gindulyte, A.; Shoemaker, B. A.; Thiessen, P. A.; He, S.; Zhang, J. Nucleic Acids Res. 2017, 45, D955.
- (4) Kim, S. Expert Opinion on Drug Discovery 2016, 11, 843.
- (5) ChemSpider (http://www.chemspider.com) (Accessed on 2/17/2017).
- (6) Pence, H. E.; Williams, A. J. Chem. Educ. 2010, 87, 1123.
- (7) ChemSpider SyntheticPages (CSSP) (http://cssp.chemspider.com/) (Accessed on 2/17/2017).
- (8) ChEMBL (https://www.ebi.ac.uk/chembl/) (Accessed on 2/17/2017).
- (9) Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A. P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L. J.; Cibrián-Uhalte, E.; Davies, M.; Dedman, N.; Karlsson, A.; Magariños, M. P.; Overington, J. P.; Papadatos, G.; Smit, I.; Leach, A. R. Nucleic Acids Res. 2017, 45, D945.
- (10) Orange Book: Approved Drug Products with Therapeutic Equivalence Evaluations (http://www.accessdata.fda.gov/scripts/cder/ob/default.cfm) (Accessed on 2/17/2017).
- (11) DailyMed (http://dailymed.nlm.nih.gov/) (Accessed on 2/17/2017).
- (12) ChEBI (https://www.ebi.ac.uk/chebi/) (Accessed on 2/17/2017).
- (13) Hastings, J.; de Matos, P.; Dekker, A.; Ennis, M.; Harsha, B.; Kale, N.; Muthukrishnan, V.; Owen, G.; Turner, S.; Williams, M.; Steinbeck, C. Nucleic Acids Res. 2013, 41, D456.
- (14) NIST Chemistry Webbook (http://webbook.nist.gov/chemistry/) (Accessed on 2/19/2017).
- (15) Linstrom, P. J.; Mallard, W. G. J. Chem. Eng. Data 2001, 46, 1059.
- (16) DrugBank (http://www.drugbank.ca/) (Accessed on 2/19/2017).
- (17) About DrugBank (http://www.drugbank.ca/about) (Accessed on 2/19/2017).
- (18) Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A. C.; Liu, Y. F.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; Tang, A.; Gabriel, G.; Ly, C.; Adamjee, S.; Dame, Z. T.; Han, B. S.; Zhou, Y.; Wishart, D. S. Nucleic Acids Res. 2014, 42, D1091.
- (19) The Human Metabolome Database (HMDB) (http://www.hmdb.ca/) (Accessed on 2/19/2017).
- (20) About the Human Metabolome Database (HMDB) (http://www.hmdb.ca/about) (Accessed on 2/19/2017).
- (21) Wishart, D. S.; Jewison, T.; Guo, A. C.; Wilson, M.; Knox, C.; Liu, Y. F.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; Bouatra, S.; Sinelnikov, I.; Arndt, D.; Xia, J. G.; Liu, P.; Yallou, F.; Bjorndahl, T.; Perez-Pineiro, R.; Eisner, R.; Allen, F.; Neveu, V.; Greiner, R.; Scalbert, A. Nucleic Acids Res. 2013, 41, D801.
- (22) ToxNet (http://toxnet.nlm.nih.gov/) (Accessed on 2/19/2017).
- (23) Factsheet - Toxicology Data Network (TOXNET) (http://www.nlm.nih.gov/pubs/factsheets/toxnetfs.html) (Accessed on 2/19/2017).
- (24) Wexler, P. Toxicology 2001, 157, 3.
- (25) Fowler, S.; Schnall, J. G. Am. J. Nurs. 2014, 114, 61.
- (26) ChemIDplus (http://chem.sis.nlm.nih.gov/chemidplus/chemidlite.jsp) (Accessed on 2/19/2017).
- (27) Fact Sheet - ChemIDplus (http://www.nlm.nih.gov/pubs/factsheets/chemidplusfs.html) (Accessed on 2/19/2017).
- (28) Hazardous Substances Data Bank (HSDB) (http://toxnet.nlm.nih.gov/newtoxnet/hsdb.htm) (Accessed on 2/19/2017).
- (29) Fact Sheet - Hazardous Substances Data Bank (HSDB) (http://www.nlm.nih.gov/pubs/factsheets/hsdbfs.html) (Accessed on 2/19/2017).
- (30) Comparative Toxicogenomics Database (CTD) (http://toxnet.nlm.nih.gov/newtoxnet/ctd.htm) (Accessed on 2/19/2017).
- (31) Fact Sheet - Comparative Toxicogenomics Database (CTD) (http://www.nlm.nih.gov/pubs/factsheets/ctdfs.html) (Accessed on 2/19/2017).
- (32) Worldwide Protein Data Bank (wwPDB) (http://www.wwpdb.org/) (Accessed on 2/19/2017).
- (33) Protein Data Bank in Europe (PDBe) (http://www.ebi.ac.uk/pdbe/) (Accessed on 2/19/2017).
- (34) Gutmanas, A.; Alhroub, Y.; Battle, G. M.; Berrisford, J. M.; Bochet, E.; Conroy, M. J.; Dana, J. M.; Montecelo, M. A. F.; van Ginkel, G.; Gore, S. P.; Haslam, P.; Hendrickx, P. M. S.; Hirshberg, M.; Lagerstedt, I.; Mir, S.; Mukhopadhyay, A.; Oldfield, T. J.; Patwardhan, A.; Rinaldi, L.; Sahni, G.; Sanz-Garcia, E.; Sen, S.; Slowley, R. A.; Velankar, S.; Wainwright, M. E.; Kleywegt, G. J. Nucleic Acids Res. 2014, 42, D285.
- (35) Protein Data Bank Japan (PDBj) (http://pdbj.org/) (Accessed on 2/19/2017).
- (36) Kinjo, A. R.; Suzuki, H.; Yamashita, R.; Ikegawa, Y.; Kudou, T.; Igarashi, R.; Kengaku, Y.; Cho, H.; Standley, D. M.; Nakagawa, A.; Nakamura, H. Nucleic Acids Res. 2012, 40, D453.
- (37) RCSB Protein Data Bank (RCSB PDB) (http://www.rcsb.org/pdb/) (Accessed on 2/19/2017).
- (38) Rose, P. W.; Prlic, A.; Bi, C. X.; Bluhm, W. F.; Christie, C. H.; Dutta, S.; Green, R. K.; Goodsell, D. S.; Westbrook, J. D.; Woo, J.; Young, J.; Zardecki, C.; Berman, H. M.; Bourne, P. E.; Burley, S. K. Nucleic Acids Res. 2015, 43, D345.
- (39) Schnoes, A. M.; Brown, S. D.; Dodevski, I.; Babbitt, P. C. PLoS Comput. Biol. 2009, 5, e1000605.
- (40) Philippi, S.; Kohler, J. Nat. Rev. Genet. 2006, 7, 482.
Contributors
Sunghwan Kim, National Center for Biotechnology Information