1.7: Accessing PubChem through a Web Interface
- Page ID
- 144262
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)PUG
PUG stands for Power User Gateway and is an Application Program Interface (API) service PubChem offers that allows users to access data programmatically. Access to this data is done through a REST or SOAP. REST is a web service type of architecture and uses web URIs (Uniform Resource identifiers). A URI is similar to the common web URL (Uniform Resource Locater) that browsers use to find web pages, but is associated with an object that may, or may not be a webpage (a URL is a type of URI). REST can provide data in many file formats, like text, html and jpeg. SOAP (Simple Object Access Protocol) is actually a protocol that works with XML files and is typically used for organizations that need higher levels of security. Although PUG works with both SOAP and REST, this course will focus on the use of REST interfaces.
REST Architecture
REST = Representational State Transfer is a way for computers to communicate over the web, where one computer may be a database server and the other is the client. One advantage to REST interfaces is that they are built upon the internet's Hypertext Transfer Protocol (http) that web browsers use, and which most people are familiar with. In essence, they are a special type of URL that interacts with specific objects with a database. A REST request is analogous to a sentence where the noun is the object and the verb is the action. Here are some typical REST verbs
- GET - retrieve a resource/object
- POST - upload a resource/object
- PUT - update a resource/object
- DELETE - remove a resource/object
In PubChem data is stored of essentially three types, each with its own identifier; compound (CID), substance (SID) and BioAssay (AID). The following figure shows the general process where you input a name, that gets converted to an identifier, and you then perform an operation to produce the type of object you are seeking and then returns an output of the file type that you are seeking.
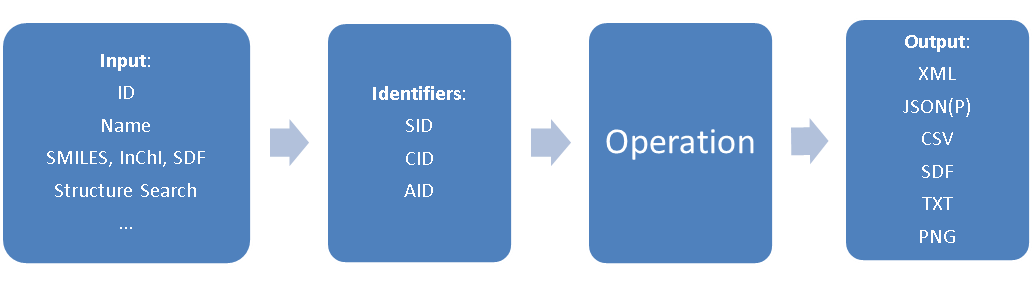
Figure \(\PageIndex{1}\): Flow chart for a REST request in PubChem (Image Credit: PubChem)
The PUG REST request is based on http (or https) and we can consider the URL to consist of four parts; the prolog, input, operation and output
|
/compound/name/aspirin |
/property/InChI |
/TXT |
|
|---|---|---|---|
|
prolog |
input |
operation |
output |
Prolog
The prolog essentially identifies the API service being used in the request.
Input
There are a variety of input methods supported
By Identifier
/substance/sid/[insert: substance ID]
/compound/cid/[insert: compound ID]
/assay/aid/[insert: Assay ID]
Examples
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/999/synonyms/txt
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/15/png
For a list of properties
For a summary of assay 999
https://pubchem.ncbi.nlm.nih.gov/rest/pug/assay/aid/999/summary/JSON
By Name
/compound/name/[insert: name of chemical]
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/glucose/PNG
By Structure
If you have a structural drawing software you can convert you image to a SMILES string or InChI Key and search with that
/compound/smiles/[insert: smiles string here]/[output]/file type
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/smiles/CC(=O)C/property/IUPACName/txt
Operation
There is a variety of data available.
Images
Images are available for all types of structure input, just finish with png
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/THC/PNG
Compound Properties
Note, these are computed properties. Actual experimental values are not available because there can be more than one value for the same property.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/THC/property/MolecularWeight/txt
The following properties can be obtained through the REST architecture
| Property | Notes |
|---|---|
|
MolecularFormula |
|
|
MolecularWeight |
The molecular weight is the sum of all atomic weights of the constituent atoms in a compound, measured in g/mol. In the absence of explicit isotope labelling, averaged natural abundance is assumed. If an atom bears an explicit isotope label, 100% isotopic purity is assumed at this location. |
|
CanonicalSMILES |
Canonical SMILES (Simplified Molecular Input Line Entry System) string. It is a unique SMILES string of a compound, generated by a “canonicalization” algorithm. |
|
IsomericSMILES |
Isomeric SMILES string. It is a SMILES string with stereochemical and isotopic specifications. |
|
InChI |
Standard IUPAC International Chemical Identifier (InChI). It does not allow for user selectable options in dealing with the stereochemistry and tautomer layers of the InChI string. |
|
InChIKey |
Hashed version of the full standard InChI, consisting of 27 characters. |
|
IUPACName |
Chemical name systematically determined according to the IUPAC nomenclatures. |
|
XLogP |
Computationally generated octanol-water partition coefficient or distribution coefficient. XLogP is used as a measure of hydrophilicity or hydrophobicity of a molecule. |
|
ExactMass |
The mass of the most likely isotopic composition for a single molecule, corresponding to the most intense ion/molecule peak in a mass spectrum. |
|
MonoisotopicMass |
The mass of a molecule, calculated using the mass of the most abundant isotope of each element. |
|
TPSA |
Topological polar surface area, computed by the algorithm described in the paper by Ertl et al. |
|
Complexity |
The molecular complexity rating of a compound, computed using the Bertz/Hendrickson/Ihlenfeldt formula. |
|
Charge |
The total (or net) charge of a molecule. |
|
HBondDonorCount |
Number of hydrogen-bond donors in the structure. |
|
HBondAcceptorCount |
Number of hydrogen-bond acceptors in the structure. |
|
RotatableBondCount |
Number of rotatable bonds. |
|
HeavyAtomCount |
Number of non-hydrogen atoms. |
|
IsotopeAtomCount |
Number of atoms with enriched isotope(s) |
|
AtomStereoCount |
Total number of atoms with tetrahedral (sp3) stereo [e.g., (R)- or (S)-configuration] |
|
DefinedAtomStereoCount |
Number of atoms with defined tetrahedral (sp3) stereo. |
|
UndefinedAtomStereoCount |
Number of atoms with undefined tetrahedral (sp3) stereo. |
|
BondStereoCount |
Total number of bonds with planar (sp2) stereo [e.g., (E)- or (Z)-configuration]. |
|
DefinedBondStereoCount |
Number of atoms with defined planar (sp2) stereo. |
|
UndefinedBondStereoCount |
Number of atoms with undefined planar (sp2) stereo. |
|
CovalentUnitCount |
Number of covalently bound units. |
|
Volume3D |
Analytic volume of the first diverse conformer (default conformer) for a compound. |
|
XStericQuadrupole3D |
The x component of the quadrupole moment (Qx) of the first diverse conformer (default conformer) for a compound. |
|
YStericQuadrupole3D |
The y component of the quadrupole moment (Qy) of the first diverse conformer (default conformer) for a compound. |
|
ZStericQuadrupole3D |
The z component of the quadrupole moment (Qz) of the first diverse conformer (default conformer) for a compound. |
|
FeatureCount3D |
Total number of 3D features (the sum of FeatureAcceptorCount3D, FeatureDonorCount3D, FeatureAnionCount3D, FeatureCationCount3D, FeatureRingCount3D and FeatureHydrophobeCount3D) |
|
FeatureAcceptorCount3D |
Number of hydrogen-bond acceptors of a conformer. |
|
FeatureDonorCount3D |
Number of hydrogen-bond donors of a conformer. |
|
FeatureAnionCount3D |
Number of anionic centers (at pH 7) of a conformer. |
|
FeatureCationCount3D |
Number of cationic centers (at pH 7) of a conformer. |
|
FeatureRingCount3D |
Number of rings of a conformer. |
|
FeatureHydrophobeCount3D |
Number of hydrophobes of a conformer. |
|
ConformerModelRMSD3D |
Conformer sampling RMSD in Å. |
|
EffectiveRotorCount3D |
Total number of 3D features (the sum of FeatureAcceptorCount3D, FeatureDonorCount3D, FeatureAnionCount3D, FeatureCationCount3D, FeatureRingCount3D and FeatureHydrophobeCount3D) |
|
ConformerCount3D |
The number of conformers in the conformer model for a compound. |
|
Fingerprint2D |
Base64-encoded PubChem Substructure Fingerprint of a molecule. |
Output
The following output formats are supported
| Output Format | Description |
|---|---|
|
XML |
standard XML, for which a schema is available |
|
JSON |
JSON, JavaScript Object Notation |
|
JSONP |
JSONP, like JSON but wrapped in a callback function |
|
ASNB |
standard binary ASN.1, NCBI’s native format in many cases |
|
ASNT |
NCBI’s human-readable text flavor of ASN.1 |
|
SDF |
chemical structure data |
|
CSV |
comma-separated values, spreadsheet compatible |
|
PNG |
standard PNG image data |
|
TXT |
plain text |
Sources
- PUG REST Tutorial https://pubchemdocs.ncbi.nlm.nih.gov/pug-rest-tutorial

