6.15: DNA Sequencing
- Page ID
- 234020
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Objectives
To briefly describe one method for DNA sequencing
We will discuss one method of reading the sequence of DNA. This method, developed by Sanger won him a second Nobel prize. To sequence a single stranded piece of DNA, the complementary strand is synthesized. Four different reaction mixtures are set up. Each contain all 4 radioactive deoxynucleotides (dATP, dCTP, dGTP, dTTP) required for the reaction and DNA polymerase. In addition, dideoxyATP (ddATP) is added to one reaction tube The dATP and ddATP attach randomly to the growing 3' end of the complementary stranded. If ddATP is added no further nucleotides can be added after since its 3' end has an H and not a OH. That's why they call it dideoxy. The new chain is terminated.. If dATP is added, the chain will continue to grow until another A needs to be added. Hence a whole series of discreet fragments of DNA chains will be made, all terminated when ddATP was added. The same scenario occurs for the other 3 tubes, which contain dCTP and ddCTP, dTTP and ddTTP, and dGTP and ddGTP respectively. All the fragments made in each tube will be placed in separate lanes for electrophoresis, where the fragments will separate by size.
Didexoynucleotides
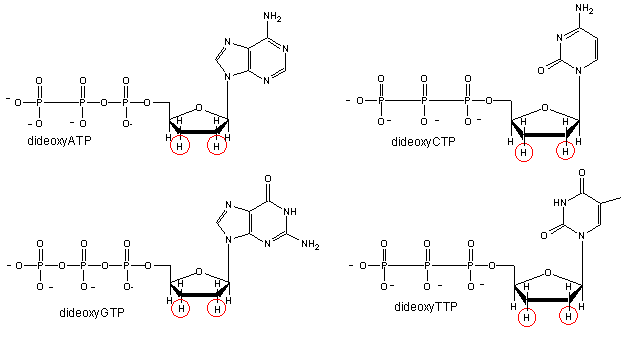
Figure: Didexoynucleotides
Example 28.6.1
You will pretend to sequence a single stranded piece of DNA as shown below. The new nucleotides are added by the enzyme DNA polymerase to the primer, GACT, in the 5' to 3' direction. You will set up 4 reaction tubes, Each tube contains all the dXTP's. In addition, add ddATP to tube 1, ddTTP to tube 2, ddCTP to tube 3, and ddGTP to tube 4. For each separate reaction mixture, determine all the possible sequences made by writing the possible sequences on one of the unfinished complementary sequences below. Cut the completed sequences from the page, determine the size of the polynucleotide sequences made, and place them as they would migrate (based on size) in the appropriate lane of a imaginary gel which you have drawn on a piece of paper. Lane 1 will contain the nucleotides made in tube 1, etc. Then draw lines under the positions of the cutout nucleotides to represent DNA bands in the gel. Read the sequence of the complementary DNA synthesized. Then write the sequence of the ssDNA that was to be sequenced.
- 5' T C A A C G A T C T G A 3' (STAND TO SEQUENCE)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
- 3' G A C T 5' (primer)
Since the DNA fragments have no detectable color, they can not be directly visualized in the gel. Alternative methods are used. In the one described above, radiolabeled ddXTP's where used. Once the sequencing gel is run, it can be dried and the bands visualized by radioautography (also called autoradiography). A place of x-ray film is placed over the dried gel in a dark environment. The radiolabeled bands will emit radiation which will expose the x-ray film directly over the bands. The film can be developed to detect the bands. In a newer technique, the primer can be labeled with a flourescent dye. If a different dye is used for each reaction mixture, all the reaction mixtures can be run in one lane of a gel. (Actually only one reaction mix containing all the ddXTP's together need be performed.) The gel can then be scanned by a laser, which detects fluorescence from the dyes, each at a different wavelength.
Figure: DNA sequencing using different fluorescent primers for each ddXTP reaction
One recent advance in sequencing allows for real-time determination of a sequence. The four deoxynucleotides are each labeled with a different fluorphore on the 5' phosphate (not the base as above). A tethered DNA polymerase elongates the DNA on a template, releasing the fluorophore into solution (i.e. the fluorophore is not incorporated into the DNA chain). The reaction takes place in a visualization chamber called a zero mode waveguide which is a cylindrical metallic chamber with a width of 70 nm and a volume of 20 zeptoliters (20 x 10-21 L). It sits on a glass support through which laser illumination of the sample is achieved. Given the small volume, non-incorporated fluorescently tagged deoxynucleotides diffuse in and out in the microsecond timescale. When a deoxynucleotide is incorporated into the DNA, its residence time is in the millisecond time scale. This allows for prolonged detection of fluorescence which give a high signal to noise ratio. This method might bring the cost of sequencing the human genome down from the initial billion dollar range to $100.