10.7: The Effect of pH on Enzyme Kinetics
- Page ID
- 41367
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)In the same way that every enzyme has an optimum temperature, so each enzyme also has an optimum pH at which it works best. For example, trypsin and pepsin are both enzymes in the digestive system which break protein chains in the food into smaller bits - either into smaller peptide chains or into individual amino acids. Pepsin works in the highly acidic conditions of the stomach. It has an optimum pH of about 1.5. On the other hand, trypsin works in the small intestine, parts of which have a pH of around 7.5. Trypsin's optimum pH is about 8.
| Enzyme | Optimal pH | Enzyme | Optimal pH |
|---|---|---|---|
| Lipase (pancreas) | 8.0 | Invertase | 4.5 |
| Lipase (stomach) | 4.0 - 5.0 | Maltase | 6.1 - 6.8 |
| Lipase (castor oil) | 4.7 | Amylase (pancreas) | 6.7 - 7.0 |
| Pepsin | 1.5 - 1.6 | Amylase (malt) | 4.6 - 5.2 |
| Trypsin | 7.8 - 8.7 | Catalase | 7.0 |
| Urease | 7.0 |
If you think about the structure of an enzyme molecule, and the sorts of bonds that it may form with its substrate, it isn't surprising that pH should matter. Suppose an enzyme has an optimum pH around 7. Imagine that at a pH of around 7, a substrate attaches itself to the enzyme via two ionic bonds. In the diagram below, the groups allowing ionic bonding are caused by the transfer of a hydrogen ion from a -COOH group in the side chain of one amino acid residue to an -NH2 group in the side chain of another.
In this simplified example, that is equally true in both the substrate and the enzyme.
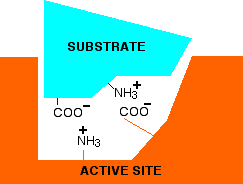
Now think about what happens at a lower pH - in other words under acidic conditions. It won't affect the -NH3+ group, but the -COO- will pick up a hydrogen ion. What you will have will be this:
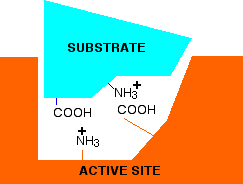
You no longer have the ability to form ionic bonds between the substrate and the enzyme. If those bonds were necessary to attach the substrate and activate it in some way, then at this lower pH, the enzyme won't work. What if you have a pH higher than 7 - in other words under alkaline conditions. This time, the -COO- group won't be affected, but the -NH3+ group will lose a hydrogen ion. That leaves . . .

Again, there is no possibility of forming ionic bonds, and so the enzyme probably won't work this time either. At extreme pH's, something more drastic can happen. Remember that the tertiary structure of the protein is in part held together by ionic bonds just like those we've looked at between the enzyme and its substrate. At very high or very low pH's, these bonds within the enzyme can be disrupted, and it can lose its shape. If it loses its shape, the active site will probably be lost completely. This is essentially the same as denaturing the protein by heating it too much.
Kinetics
The rates of enzyme-catalysed reactions vary with pH and often pass through a maximum as the pH is varied. If the enzyme obeys Michaelis-Menten kinetics the kinetic parameters k0 and kA often behave similarly. The pH at which the rate or a suitable parameter is a maximum is called the pH optimum and the plot of rate or parameter against pH is called a pH profile. Neither the pH optimum nor the pH profile of an enzyme has any absolute significance and both may vary according to which parameter is plotted and according to the conditions of the measurements.
If the pH is changed and then brought back to its original value, the behavior is said to be reversible if the original properties of the enzyme are restored; otherwise it is irreversible. Reversible pH behavior may occur over a narrow range of pH, but effects of large changes in pH are in most cases irreversible. The diminution in rate as the pH is taken to the acid side of the optimum can be regarded as inhibition by hydrogen ions. The diminution in rate on the alkaline side can be regarded as inhibition by hydroxide ions. The equations describing pH effects are therefore analogous to inhibition equations. For single-substrate reactions the pH behavior of the parameters k0 and kA can sometimes be represented by an equation of the form
\[ k = \dfrac{k_{opt}}{1 + \dfrac{[H^+]}{K_1} + \dfrac{K_2}{[H^+]}} \label{eq1}\]
in which k represents k0 or kA, and \(k_{opt}\) is the value of the same parameter that would be observed if the enzyme existed entirely in the optimal state of protonation; it may be called the pH-independent value of the parameter. The constants K1 and K2 can sometimes be identified as acid dissociation constants for the enzyme. substrates or other species in the reaction mixture. The identification is, however, never straight forward and has to be justified by independent evidence. The behavior is frequently much more complicated than represented by Equation \(\ref{eq1}\).
It is not accidental that this section has referred exclusively to pH dependences of k0 and kA. The pH dependence of the initial rate or, worse, the extent of reaction after a given time is rarely meaningful; the pH dependence of the Michaelis constant is often too complex to be readily interpretable.
The pH dependence of the Michaelis constant is often too complex to be readily interpretable.
Quenching Enzyme Activity
In principle, we can use any of the range of possible analytical techniques to follow a reaction’s kinetics provided that the reaction does not proceed to any appreciable extent during the time it takes to make a measurement. As you might expect, this requirement places a serious limitation on kinetic methods of analysis. If the reaction’s kinetics are slow relative to the analysis time, then we can make our measurements without the analyte undergoing a significant change in concentration. When the reaction’s rate is too fast—which often is the case—then we introduce a significant error if our analysis time is too long.
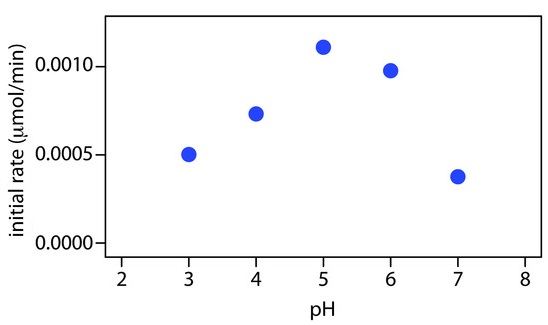
One solution to this problem is to stop, or quench the reaction by adjusting experimental conditions. For example, many reactions show a strong pH dependency, and may be quenched by adding a strong acid or a strong base. Figure 13.7 shows a typical example for the enzymatic analysis of p-nitrophenylphosphate using the enzyme wheat germ acid phosphatase to hydrolyze the analyte to p-nitrophenol.
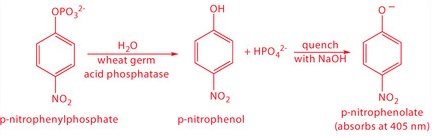
The reaction has a maximum rate at a pH of 5. Increasing the pH by adding NaOH quenches the reaction and converts the colorless p-nitrophenol to the yellow-colored p-nitrophenolate, which absorbs at 405 nm.