16.5: Structure and Function of Proteins
- Page ID
- 153898
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Describe the four levels of protein structure.
- Identify the types of attractive interactions that hold proteins in their most stable three-dimensional structure
- Explain the role of an enzyme in the body.
Each of the thousands of naturally occurring proteins has its own characteristic amino acid composition and sequence that result in a unique three-dimensional shape. Since the 1950s, scientists have determined the amino acid sequences and three-dimensional conformation of numerous proteins and thus obtained important clues on how each protein performs its specific function in the body.
Proteins are compounds of high molar mass consisting largely or entirely of chains of amino acids. Because of their great complexity, protein molecules cannot be classified on the basis of specific structural similarities, as carbohydrates and lipids are categorized. The two major structural classifications of proteins are based on far more general qualities: whether the protein is (1) fiberlike and insoluble or (2) globular and soluble. Some proteins, such as those that compose hair, skin, muscles, and connective tissue, are fiberlike. These fibrous proteins are insoluble in water and usually serve structural, connective, and protective functions. Examples of fibrous proteins are keratins, collagens, myosins, and elastins. Hair and the outer layer of skin are composed of keratin. Connective tissues contain collagen. Myosins are muscle proteins and are capable of contraction and extension. Elastins are found in ligaments and the elastic tissue of artery walls.
Globular proteins, the other major class, are soluble in aqueous media. In these proteins, the chains are folded so that the molecule as a whole is roughly spherical. Familiar examples include egg albumin from egg whites and serum albumin in blood. Serum albumin plays a major role in transporting fatty acids and maintaining a proper balance of osmotic pressures in the body. Hemoglobin and myoglobin, which are important for binding oxygen, are also globular proteins.
Levels of Protein Structure
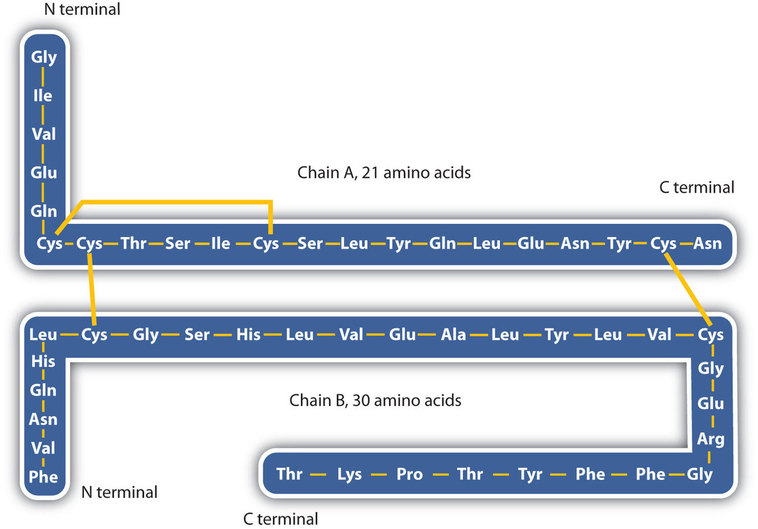
The structure of proteins is generally described as having four organizational levels. The first of these is the primary structure, which is the number and sequence of amino acids in a protein’s polypeptide chain or chains, beginning with the free amino group and maintained by the peptide bonds connecting each amino acid to the next. The primary structure of insulin, composed of 51 amino acids, is shown in Figure \(\PageIndex{1}\).
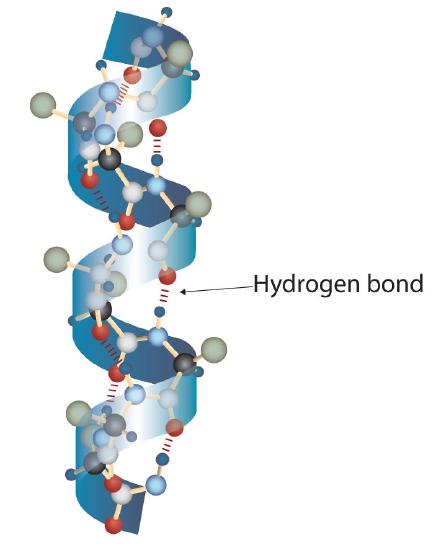
A protein molecule is not a random tangle of polypeptide chains. Instead, the chains are arranged in unique but specific conformations. The term secondary structure refers to the fixed arrangement of the polypeptide backbone. On the basis of X ray studies, Linus Pauling and Robert Corey postulated that certain proteins or portions of proteins twist into a spiral or a helix. This helix is stabilized by intrachain hydrogen bonding between the carbonyl oxygen atom of one amino acid and the amide hydrogen atom four amino acids up the chain (located on the next turn of the helix) and is known as a right-handed α-helix. X ray data indicate that this helix makes one turn for every 3.6 amino acids, and the side chains of these amino acids project outward from the coiled backbone (Figure \(\PageIndex{2}\)). The α-keratins, found in hair and wool, are exclusively α-helical in conformation. Some proteins, such as gamma globulin, chymotrypsin, and cytochrome c, have little or no helical structure. Others, such as hemoglobin and myoglobin, are helical in certain regions but not in others.
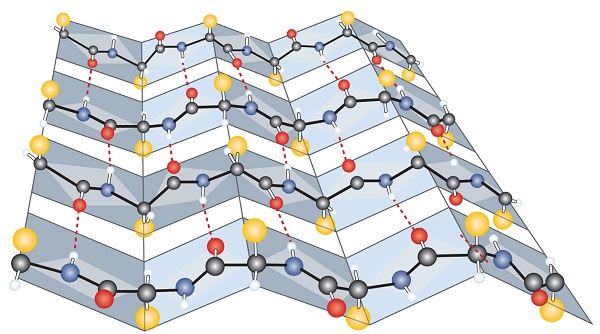
Another common type of secondary structure, called the β-pleated sheet conformation, is a sheetlike arrangement in which two or more extended polypeptide chains (or separate regions on the same chain) are aligned side by side. The aligned segments can run either parallel or antiparallel—that is, the N-terminals can face in the same direction on adjacent chains or in different directions—and are connected by interchain hydrogen bonding (Figure \(\PageIndex{3}\)). The β-pleated sheet is particularly important in structural proteins, such as silk fibroin. It is also seen in portions of many enzymes, such as carboxypeptidase A and lysozyme.
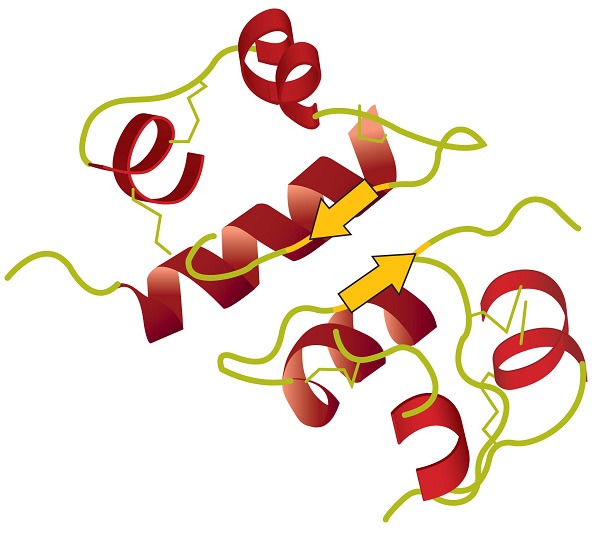
Tertiary structure refers to the unique three-dimensional shape of the protein as a whole, which results from the folding and bending of the protein backbone. The tertiary structure is intimately tied to the proper biochemical functioning of the protein. Figure \(\PageIndex{4}\) shows a depiction of the three-dimensional structure of insulin.
Four major types of attractive interactions determine the shape and stability of the tertiary structure of proteins.
You studied several of them previously.
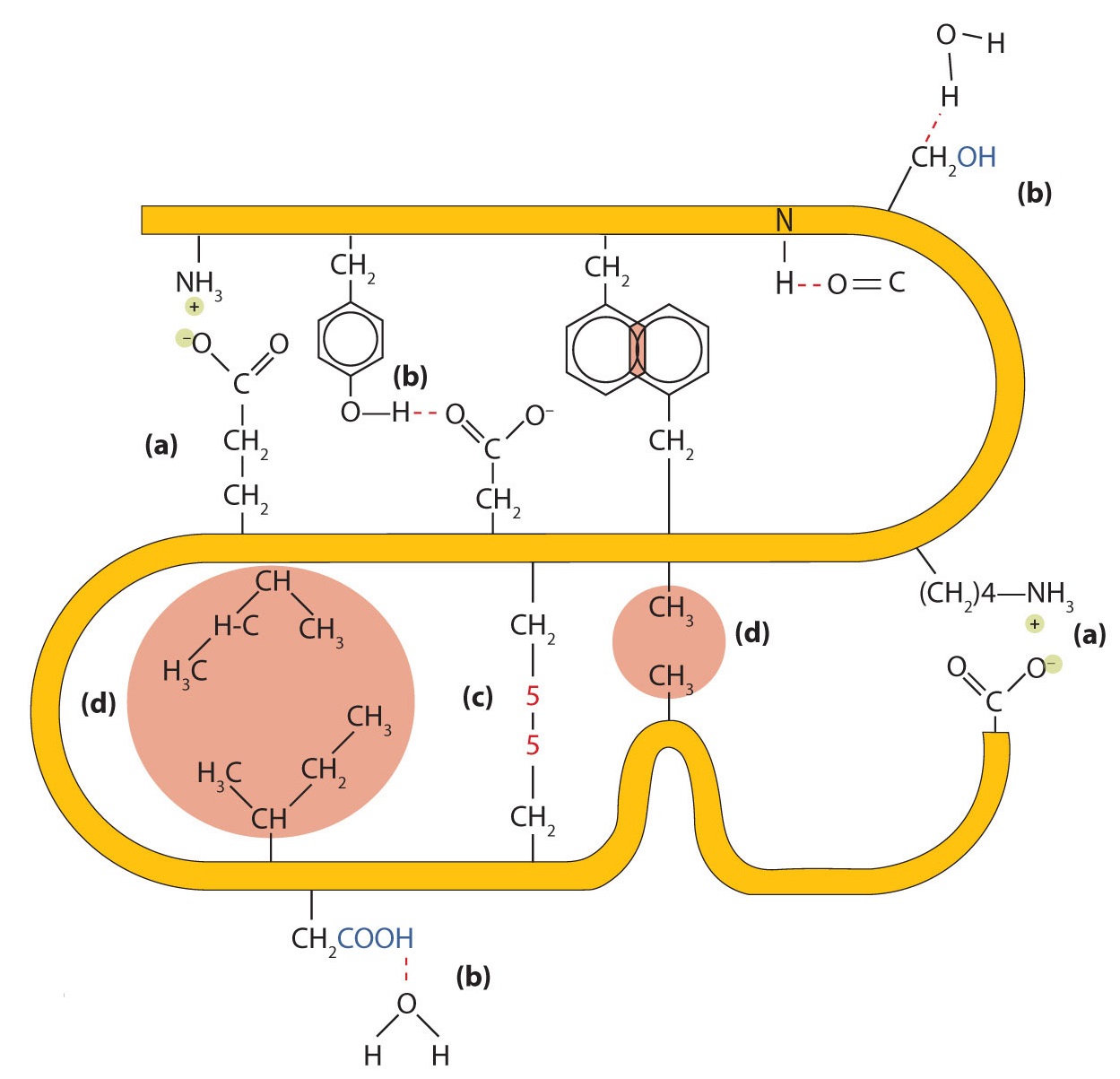
- Ionic bonding. Ionic bonds result from electrostatic attractions between positively and negatively charged side chains of amino acids. For example, the mutual attraction between an aspartic acid carboxylate ion and a lysine ammonium ion helps to maintain a particular folded area of a protein (part (a) of Figure \(\PageIndex{5}\)).
- Hydrogen bonding. Hydrogen bonding forms between a highly electronegative oxygen atom or a nitrogen atom and a hydrogen atom attached to another oxygen atom or a nitrogen atom, such as those found in polar amino acid side chains. Hydrogen bonding (as well as ionic attractions) is extremely important in both the intra- and intermolecular interactions of proteins (part (b) of Figure \(\PageIndex{5}\)).
- Disulfide linkages. Two cysteine amino acid units may be brought close together as the protein molecule folds. Subsequent oxidation and linkage of the sulfur atoms in the highly reactive sulfhydryl (SH) groups leads to the formation of cystine (part (c) of Figure \(\PageIndex{5}\)). Intrachain disulfide linkages are found in many proteins, including insulin (yellow bars in Figure \(\PageIndex{1}\)) and have a strong stabilizing effect on the tertiary structure.
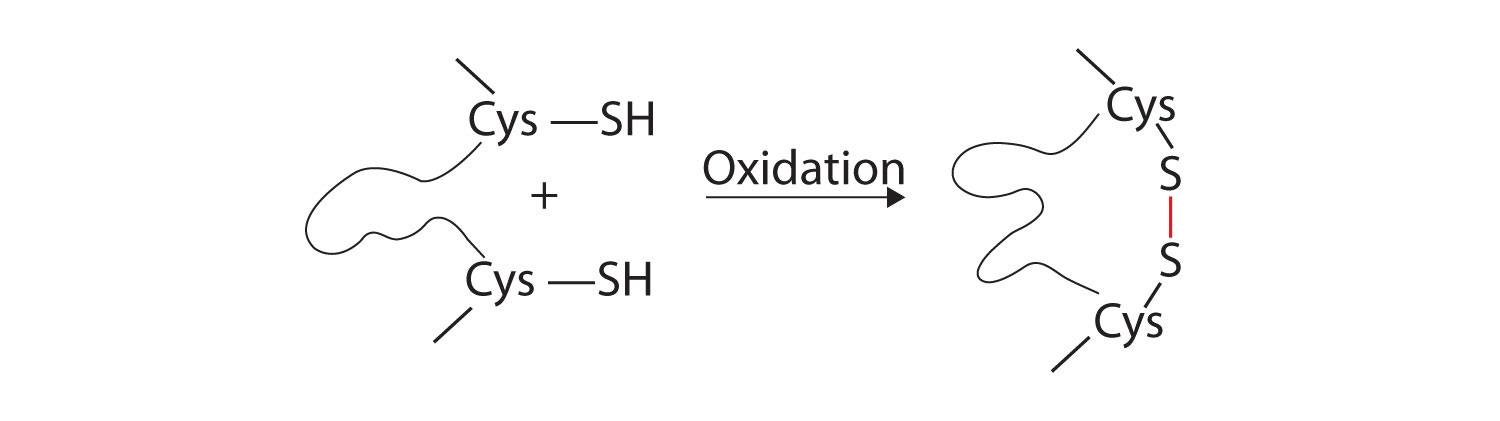
- Dispersion forces. Dispersion forces arise when a normally nonpolar atom becomes momentarily polar due to an uneven distribution of electrons, leading to an instantaneous dipole that induces a shift of electrons in a neighboring nonpolar atom. Dispersion forces are weak but can be important when other types of interactions are either missing or minimal (part (d) of Figure \(\PageIndex{5}\)). This is the case with fibroin, the major protein in silk, in which a high proportion of amino acids in the protein have nonpolar side chains. The term hydrophobic interaction is often misused as a synonym for dispersion forces. Hydrophobic interactions arise because water molecules engage in hydrogen bonding with other water molecules (or groups in proteins capable of hydrogen bonding). Because nonpolar groups cannot engage in hydrogen bonding, the protein folds in such a way that these groups are buried in the interior part of the protein structure, minimizing their contact with water.
When a protein contains more than one polypeptide chain, each chain is called a subunit. The arrangement of multiple subunits represents a fourth level of structure, the quaternary structure of a protein. Hemoglobin, with four polypeptide chains or subunits, is the most frequently cited example of a protein having quaternary structure (Figure \(\PageIndex{6}\)). The quaternary structure of a protein is produced and stabilized by the same kinds of interactions that produce and maintain the tertiary structure. A schematic representation of the four levels of protein structure is in Figure \(\PageIndex{7}\).
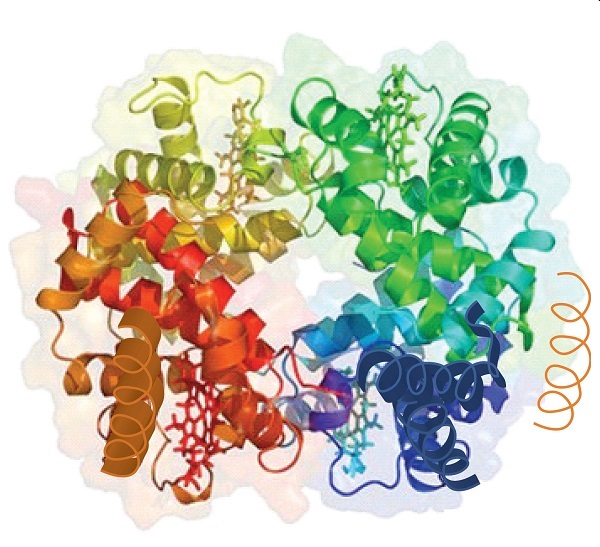
Source: Image from the RCSB PDB (www.pdb.org) of PDB ID 1I3D (R.D. Kidd, H.M. Baker, A.J. Mathews, T. Brittain, E.N. Baker (2001) Oligomerization and ligand binding in a homotetrameric hemoglobin: two high-resolution crystal structures of hemoglobin Bart's (gamma(4)), a marker for alpha-thalassemia. Protein Sci. 1739–1749).
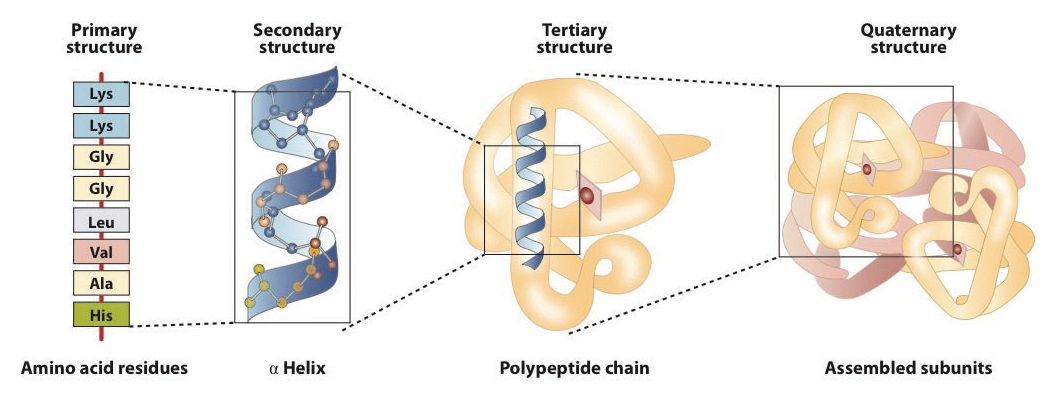
The primary structure consists of the specific amino acid sequence. The resulting peptide chain can twist into an α-helix, which is one type of secondary structure. This helical segment is incorporated into the tertiary structure of the folded polypeptide chain. The single polypeptide chain is a subunit that constitutes the quaternary structure of a protein, such as hemoglobin that has four polypeptide chains.
Globular and Fibrous Proteins
Once proteins form and have developed all levels of their structure, they can be classified as either fibrous or globular. These classifications give the basic shape of the entire protein molecule. While many proteins are globular proteins (see figure below), keratin proteins are fibrous (see figure below) and make up the hair, nails, and the outer layer of skin.
Enzymes: Exquisite Precision Machines
The first enzyme to be isolated was discovered in 1926 by American chemist James Sumner, who crystallized the protein. The enzyme was urease, which catalyzes the hydrolytic decomposition of urea, a component of urine, into ammonia and carbon dioxide.
\[\ce{H_2NCON_2} \left( aq \right) + \ce{H_2O} \left( l \right) \overset{\text{urease}}{\rightarrow} 2 \ce{NH_3} \left( g \right) + \ce{CO_2} \left( g \right) \nonumber \]
His discovery was ridiculed at first because nobody believed that enzymes would behave the same way that other chemicals did. Sumner was eventually proven right and won the Nobel Prize in Chemistry in 1946.
Enzymes and Biochemical Reactions
Most chemical reactions within organisms would be impossible under the conditions in cells. For example, the body temperature of most organisms is too low for reactions to occur quickly enough to carry out life processes. Reactants may also be present in such low concentrations that it is unlikely they will meet and collide. Therefore, the rate of most biochemical reactions must be increased by a catalyst. A catalyst is a chemical that speeds up chemical reactions. In organisms, catalysts are called enzymes. Essentially, enzymes are biological catalysts.
Like other catalysts, enzymes are not reactants in the reactions they control. They help the reactants interact but are not used up in the reactions. Instead, they may be used over and over again. Unlike other catalysts, enzymes are usually highly specific for particular chemical reactions. They generally catalyze only one or a few types of reactions.
Enzymes are extremely efficient in speeding up reactions. They can catalyze up to several million reactions per second. As a result, the difference in rates of biochemical reactions with and without enzymes may be enormous. A typical biochemical reaction might take hours or even days to occur under normal cellular conditions without an enzyme, but less than a second with an enzyme.
Figure \(\PageIndex{10}\) diagrams a typical enzymatic reaction. A substrate is the molecule or molecules on which the enzyme acts. In the urease catalyzed reaction, urea is the substrate.
The first step in the reaction is that the substrate binds to a specific part of the enzyme molecule, known as the active site. The binding of the substrate is dictated by the shape of each molecule. Side chains on the enzyme interact with the substrate in a specific way, resulting in the making and breaking of bonds. The active site is the place on an enzyme where the substrate binds. An enzyme folds in such a way that it typically has one active site, usually a pocket or crevice formed by the folding pattern of the protein. Because the active site of an enzyme has such a unique shape, only one particular substrate is capable of binding to that enzyme. In other words, each enzyme catalyzes only one chemical reaction with only one substrate. Once the enzyme/substrate complex is formed, the reaction occurs and the substrate is transformed into products. Finally, the product molecule or molecules are released from the active site. Note that the enzyme is left unaffected by the reaction and is now capable of catalyzing the reaction of another substrate molecule.
For many enzymes, the active site follows a lock and key (Figure \(\PageIndex{11A}\) ) model where the substrate fits exactly into the active site. The enzyme and substrate must be a perfect match so the enzyme only functions as a catalyst for one reaction. Other enzymes have an induced fit (Figure \(\PageIndex{11B}\) ) model. In an induced fit model, the active site can make minor adjustments to accommodate the substrate. This results in an enzyme that is capable of interacting with a small group of similar substrates. Look at the shape of the active site compared to the shape of the substrate in B of the figure below. The active site adjusts to accommodate the substrate.
Inhibitors
An inhibitor is a molecule which interferes with the function of an enzyme, either by slowing or stopping the chemical reaction. Inhibitors can work in a variety of ways, but one of the most common is illustrated in the figure below.
A competitive inhibitor binds competitively at the active site and blocks the substrate from binding. Since no reaction occurs with the inhibitor, the enzyme is prevented from catalyzing the reaction.
A non-competitive inhibitor does not bind at the active site. It attaches at an allosteric site, which is some other site on the enzyme, and changes the shape of the protein. The allosteric site is any site on the enzyme that is not the active site. The attachment of the non-competitive inhibitor to the allosteric site results in a shift in three-dimensional structure that alters the shape of the active site so that the substrate will no longer fit in the active site properly (see figure below).
Cofactors and Coenzymes
Some enzymes require the presence of another substrate as a "helper" molecule in order to function properly. Cofactors and coenzymes serve in this role. Cofactors are inorganic species and coenzymes are small organic molecules. Many vitamins, such as B vitamins, are coenzymes. Some metal ions which function as cofactors for various enzymes include zinc, magnesium, potassium, and iron.
Catalytic Activity of Enzymes
Enzymes generally lower activation energy by reducing the energy needed for reactants to come together and react. One way that enzymes act is to bring reactants (substrates) together so they don't have to expend energy moving about until they collide at random. Enzymes bind both reactant molecules (substrates), tightly and specifically, at a site on the enzyme's active site. Enzymes can also bring molecules to the active site to break them apart. For example, sucrase is the enzyme for the breakdown of sucrose which enters the active site of the enzyme and helps weaken the interactions between the fructose and glucose that make up sucrose. Sucrase is specific to the breakdown of sucrose as are most enzymes. The active site is specific for the reactants of the biochemical reaction the enzyme catalyzes. Similar to puzzle pieces fitting together, the active site can only bind certain substrates. The activities of enzymes also depend on the temperature, concentration, and the pH of the surroudings.
Concentration
As with most reactions, the concentration of the reactant(s) affects the reaction rate. This is also true in enzyme concentration. When either substrate or enzyme concentration is low, the rate of the reaction will be slower than where there are higher concentrations. The two species must interact for a reaction to occur and higher concentrations of one or both will result in more effective interactions between the two.
However, continuing to increase the substrate's concentration will not always increase the reaction rate. This is because at some point, all of the enzymes will be occupied and unavailable to bind with another substrate molecule until the substrate forms a product molecule and is released from the enzyme.
pH
Some enzymes work best at acidic pHs, while others work best in neutral environments. For example, digestive enzymes secreted in the acidic environment (low pH) of the stomach help break down proteins into smaller molecules. The main digestive enzyme in the stomach is pepsin, which works best at a pH of about 1.5. These enzymes would not work optimally at other pHs. Trypsin is another enzyme in the digestive system, which breaks protein chains in food into smaller particles. Trypsin works in the small intestine, which is not an acidic environment. Trypsin's optimum pH is about 8.
Different reactions and different enzymes will achieve their maximum rate at certain pH values. As shown in the figure below, the enzyme achieves a maximum reaction rate at a pH of 4. Notice that the reaction will continue at lower and higher pH values because the enzyme will still function at other pH values but will not be as effective. At very high or very low pH values, denaturation will occur because an enzyme is just a protein with a specific function.
Temperature
As with pH, reactions also have an ideal temperature where the enzyme functions most effectively. It will still function at higher and lower temperatures, but the rate will be less. For many biological reactions, the ideal temperature is at physiological conditions which is around \(37^\text{o} \text{C}\) which is normal body temperature. Many enzymes lose function at lower and higher temperatures. At higher temperatures, an enzyme's shape deteriorates. Only when the temperature comes back to normal does the enzyme regain its shape and normal activity unless the temperature was so high that it caused irreversible damage.
Applications of Enzymes
Enzymes are used in the and other industrial applications when extremely specific catalysts are required (Table \(\PageIndex{1}\)). Enzymes in general are limited in the number of reactions they have evolved to catalyze and also by their lack of stability in and at high temperatures. As a consequence, is an active area of research and involves attempts to create new enzymes with novel properties, either through rational design or in vitro evolution. These efforts have begun to be successful, and a few enzymes have now been designed "from scratch" to catalyze reactions that do not occur in nature.
| Application | Enzymes used | Uses |
|---|---|---|
| Biofuel industry | Cellulases | Break down cellulose into sugars that can be fermented to produce cellulosic ethanol. |
| Ligninases | Pretreatment of biomass for biofuel production. | |
| Biological detergent | Proteases, amylases, lipases | Remove protein, starch, and fat or oil stains from laundry and dish ware. |
| Mannanases | Remove food stains from the common food additive guar gum. | |
| Brewing industry | Amylase, glucanases, proteases | Split polysaccharides and proteins in the malt. |
| Betaglucanases | Improve the wort and beer filtration characteristics. | |
| Amyloglucosisdase and pullulanases | Make low-calorie beer and adjust fermentability. | |
| Acetolacatate deacrboxylase (ALDC) | Increase fermentation efficiency by reducing diacetyl formation. | |
| Culinary uses | Papain | Tenderize meat for cooking. |
| Dairy industry | Renin | Hydrolyze protein in the manufacture of cheese. |
| Lipases | Produce Camembert cheese and blue cheese such as Roquefort. | |
| Food processing | Amylases | Produce sugars from starch, such as in making high-fructose corn syrup. |
| Proteases | Lower the protein level of flour, as in biscuit-making. | |
| Trypsin | Manufacture hypoallergenic baby foods. | |
| Cellulases, pectinases | Clarify fruit juices. | |
| Molecular biology | Nucleases, DNA ligase, and polymerases | Use restriction digestion and the polymerase chain reaction to create recombinant DNA. |
| Paper industry | Xylanases, hemicellulases, and lignin peroxidases | Remove lignin from Kraft pulp. |
| Personal care | Proteases | Remove proteins on cotact lenses to prevent infections. |
| Starch industry | Amylases | Convert starch into glucose and various syrups. |
Summary
- Proteins can be divided into two categories: fibrous, which tend to be insoluble in water, and globular, which are more soluble in water. A protein may have up to four levels of structure.
- The primary structure consists of the specific amino acid sequence.
- The resulting peptide chain can form an α-helix or β-pleated sheet (or local structures not as easily categorized), which is known as secondary structure. These segments of secondary structure are incorporated into the tertiary structure of the folded polypeptide chain.
- The quaternary structure describes the arrangements of subunits in a protein that contains more than one subunit. Four major types of attractive interactions determine the shape and stability of the folded protein: ionic bonding, hydrogen bonding, disulfide linkages, and dispersion forces.
- An enzyme is an organic catalyst produced by a living cell. Enzymes are such powerful catalysts that the reactions they promote occur rapidly at body temperature.
- The molecule or molecules on which an enzyme acts are called its substrates.
- An enzyme has an active site where its substrate or substrates bind to form an enzyme-substrate complex.
- The original lock and key model of enzyme and substrate binding pictured a rigid enzyme of unchanging configuration binding to the appropriate substrate. The newer induced fit model describes the enzyme active site as changing its conformation after binding to the substrate.
- Enzymes have numerous applications in the food and dairy industry, biofuel industry, paper industry etc.
Contributors and Attributions
- Libretext: The Basics of GOB Chemistry (Ball et al.)
Allison Soult, Ph.D. (Department of Chemistry, University of Kentucky)
- Wikipedia

