5.5: Proteins
- Page ID
- 189779
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Protein structures can be complicated. Their NMR spectra can be very complicated. For that reason, multi-dimensional NMR techniques are often used to determine protein structures. On this page, we will look at just a few of the techniques, including COSY, TOCSY and HNCA.
Note that HNCA is not an example of 2D NMR. It is an example of 3D NMR. It shows a correlation between an amide proton, the amide nitrogen to which it is attached, and the carbons that are attached to the amide nitrogen. HNCA data are viewed in slices, in which we just look at one nitrogen at a time. One axis shows the shift of the proton attached to that nitrogen, and the other axis shows the shifts of the carbons attached to the nitrogen.
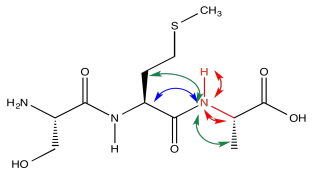
Consider a simple tripeptide.
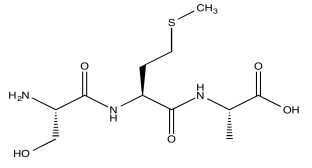
HNCA exploits a magnetization transfer, similar to the process used in a COSY or HMBC experiment. In this case, the experiment involves three different kinds of nuclei. An amide proton is irradiated (H); it transfers information to the attached nitrogen (N) and then to the alpha carbon (CA).
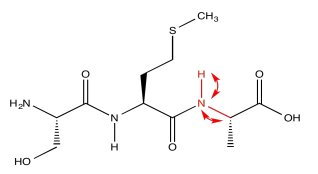
The results of the HNCA experiment display the relationship between these three nuclei. We have seen how a two-dimensional experiment can be displayed, but displaying a three-dimensional experiment would require a z axis. That might be difficult. Instead, HNCA results are displayed as "slices". A slice shows only the 13C and 1H data related to a particular 15N resonance.
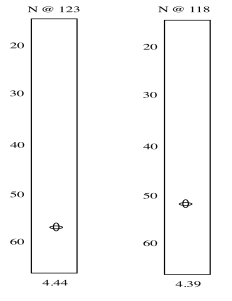
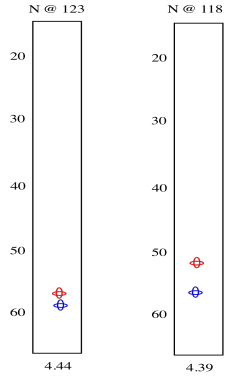
So we see there is an amide nitrogen that shows up at 123 ppm in the 15N spectrum. It is attached to a carbon that shows up at 57 ppm in the 13C spectrum and a proton that shows up at 4.44 ppm in the 1H spectrum. A second slice shows similar results for the second amide H-N-Cα group in the tripeptide.
HNCA experiments are complicated. The way the experiment is run allows different kinds of information to be obtained, much like an HMBC and HMQC show different information from fundamentally similar methods.
Running the HNCA in another way, we can actually see the alpha carbon on the other side of the carbonyl, in addition to the alpha carbon adjacent to the amide position we are looking at.
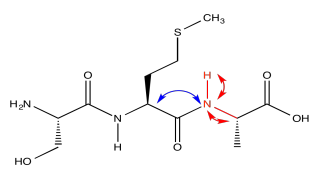
The red peaks below are the ones we had from the previous experiment. They show that relationship between the amide proton, the amide nitrogen and the alpha carbon. The blue peak shows the alpha carbon on the other side of the carbonyl. Notice that the blue peak in the slice on the right is the same as the red peak in the slice on the left. Of course, in a tripeptide, one of the amino acids is right next to the other one.
It still may not be clear which slice comes from which amide nitrogen. However, there is another variation on this experiment that can help. This one shows not only the alpha carbons on both sides of the amide nitrogen, but the beta carbons as well.
In the slices below, in addition to the red peaks and the blue peaks we had before, we now have a couple of green peaks. Those green peaks show the beta carbons on either side.
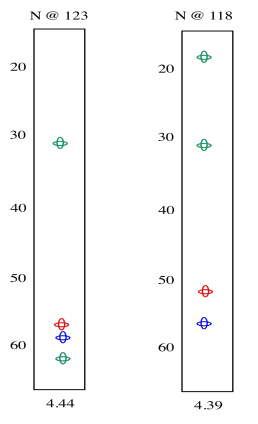
On the left slice, there is a green peak at 30 and one at 62. The peak at 62 must be the beta carbon of serine, since it is next to an oxygen. The peak at 30 is probably the beta carbon in methionine. In the right slice, the peak at 20 must be the methyl carbon in alanine. The beta carbon of methionine shows up again, because methionine is in the middle of this tripeptide -- it is next to both amide positions. Now we can see that the left slice came from the left amide position in the tripeptide, as drawn above, whereas the slice on the right came from the right amide position.
Exercise \(\PageIndex{1}\)
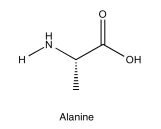
Identify the following amino acid.
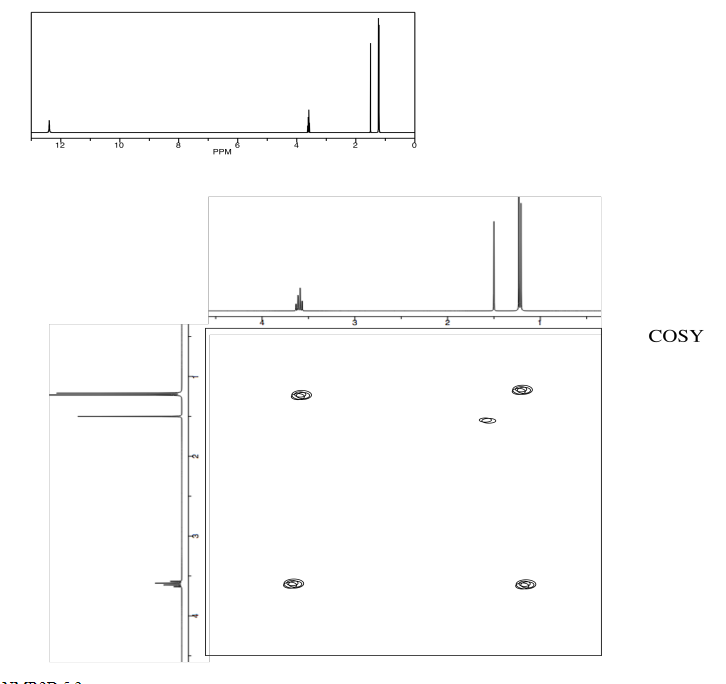
- Answer
-
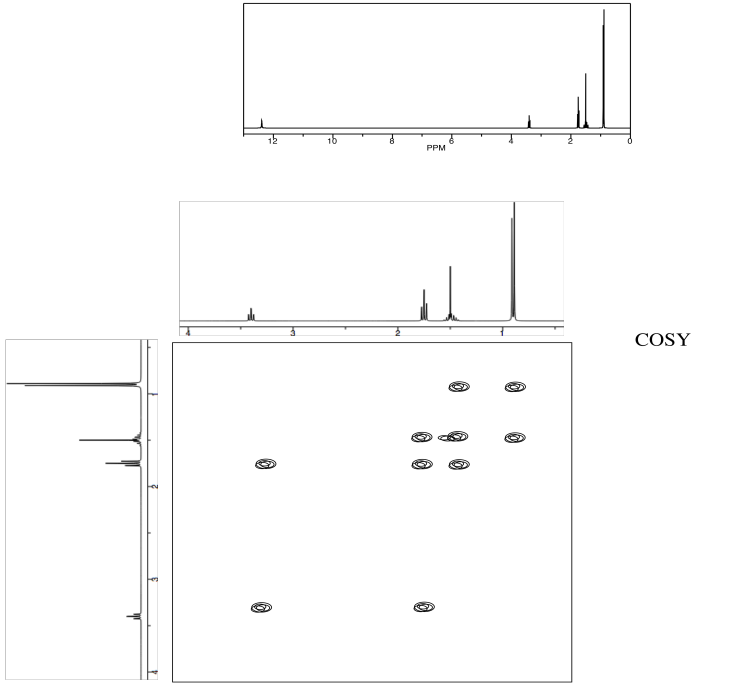
Exercise \(\PageIndex{2}\)
Identify the following amino acid.
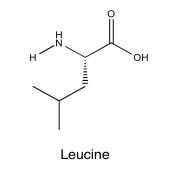
- Answer
-
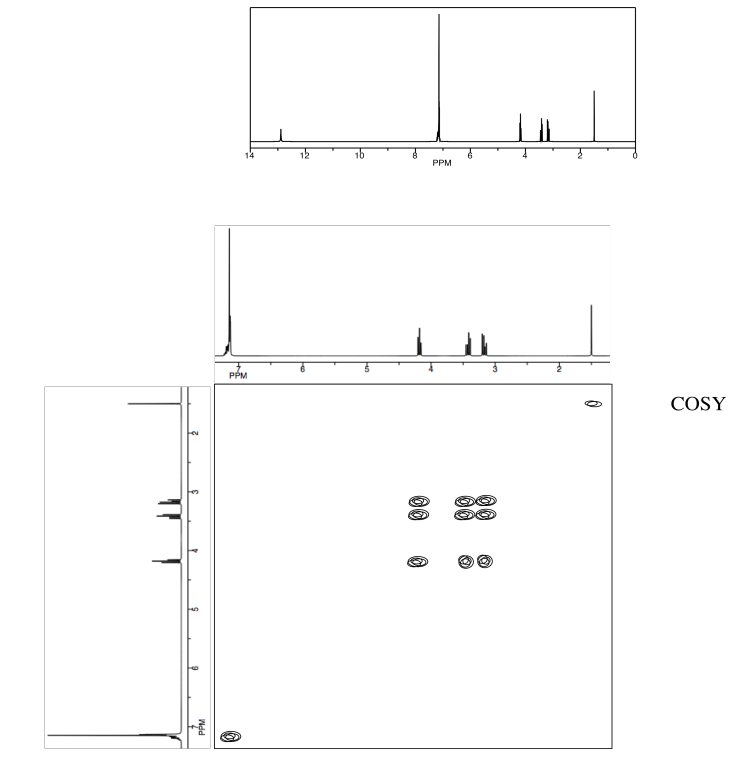
Exercise \(\PageIndex{3}\)
Identify the following amino acid.
- Answer
-
Exercise \(\PageIndex{4}\)
Identify this dipeptide.
- Use the TOCSY to identify the two amino acids involved.
- Use HNCA to determine the connectivity.
Useful References
- TOCSY/COSY of amino acids: http://www.bp.uni-bayreuth.de/NMR/nmr_aminotocsy.html
- TOCSY of all AA: http://www.bp.uni-bayreuth.de/NMR/nmr_alltocsy.html
- Standard Shifts for AA: http://www.bmrb.wisc.edu/ref_info/statsel.htm
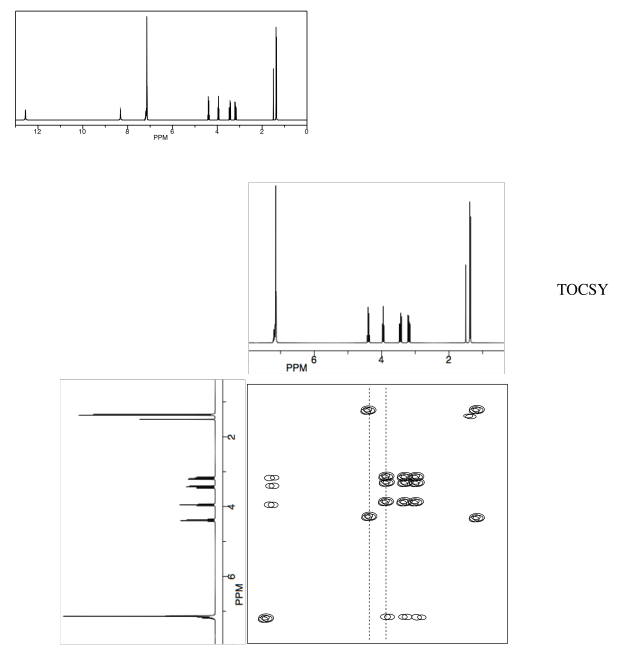
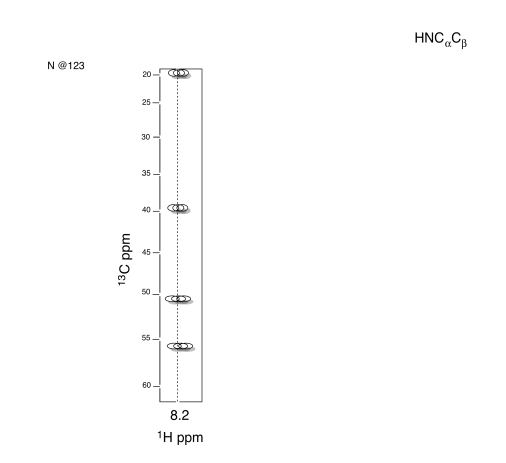
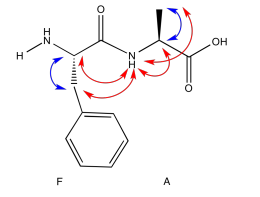
- Answer
-
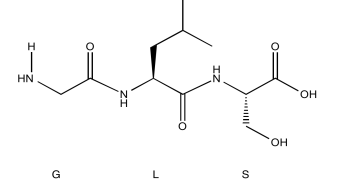
Exercise \(\PageIndex{5}\)
Identify this tripeptide.
- Use the TOCSY to identify the two amino acids involved.
- Use HNCA to determine the connectivity.
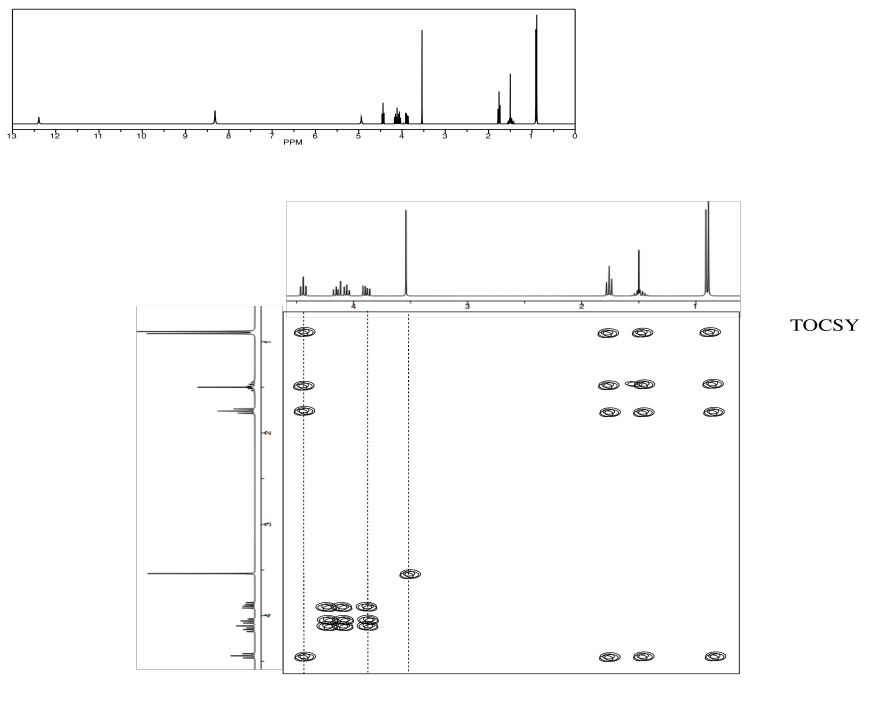
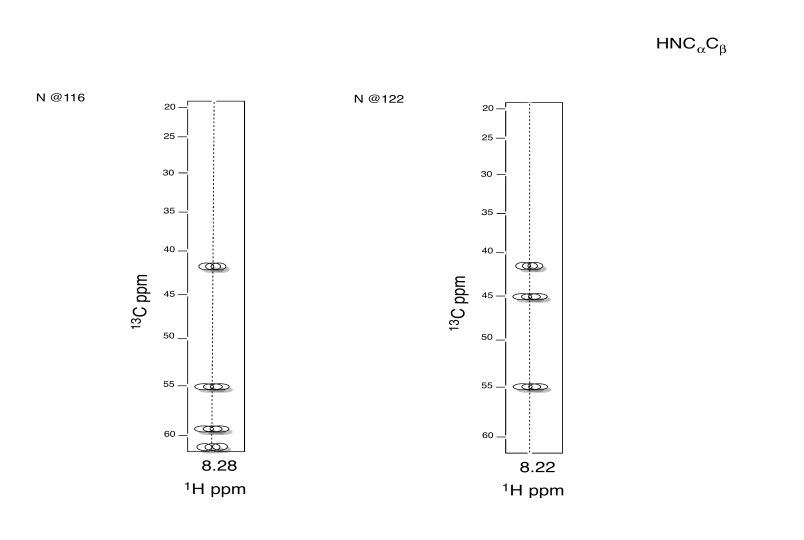
- Answer
-
Sources
1H NMR, TOCSY, HNCA and COSY spectra simulated.


