20: Protein Folding
- Page ID
- 294355
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Composed of 50–500 amino acids linked in 1D sequence by the polypeptide backbone
- The amino acid physical and chemical properties of the 20 amino acids dictate an intricate and functional 3D structure.
- Folded structure is energetic ground state (Anfinsen)

Many proteins spontaneously refold into native form in vitro with high fidelity and high speed.
Different approaches to studying this phenomenon:
- How does the primary sequence encode the 3D structure?
- Can you predict the 3D fold from a primary sequence?
- Design a polypeptide chain that folds into a known structure.
- What is the mechanism by which a disordered chain rapidly adopts its native structure?
Our emphasis here is mechanistic. What drives this process? The physical properties of the connected pendant chains interacting cooperatively give rise to the structure.
It is said that the primary sequence dictates the three-dimensional structure, but this is not the whole story, and it emphasizes a certain perspective. Certainly we need water, and defined thermodynamic conditions in temperature, pH, and ionic strength. In a sense the protein is the framework and the solvent is the glue. Folded proteins may not be as structured from crystal structures, as one is led to believe.
Kinetics and Dynamics
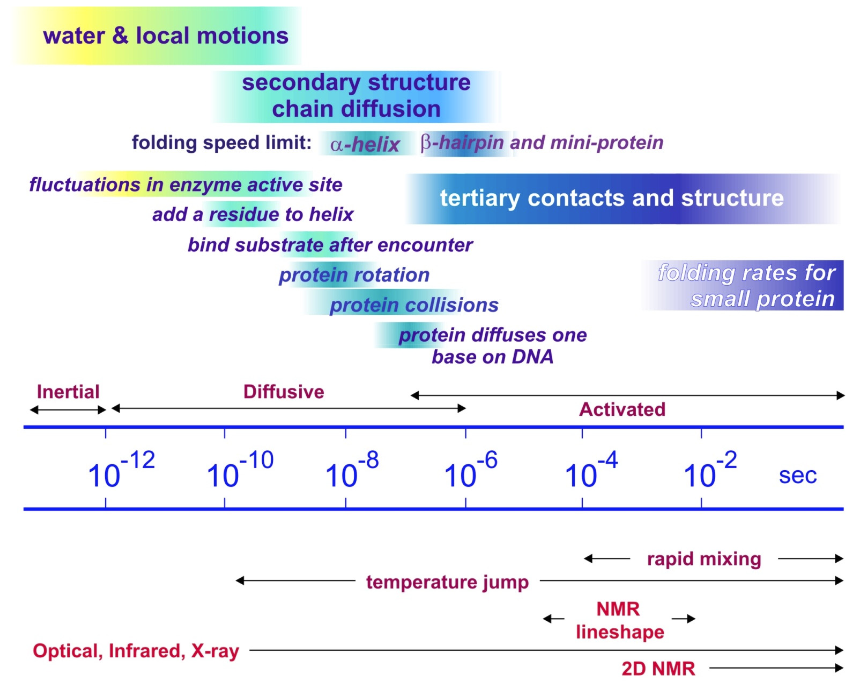
Observed protein folding time scales span decades. Observations for protein folding typically measured in ms, seconds, and minutes. This is the time scale for activated folding across a free-energy barrier. The intrinsic time scale for the underlying diffusive processes that allow conformations to evolve and local contacts to be formed through free diffusion is ps to μs. The folding of small secondary structure happens on 0.1–1 μs for helices and ~1–10 μs for hairpins. The fastest folding mini-proteins (20–30 residues) is ~1 μs.
Cooperativity
What drives this? Some hints:
Levinthal’s paradox1
The folded configuration cannot be found through a purely random search process.
- Assume: o3 states/amino acid linkage o100 linkages
- 3100 = 5 x 1047 states oSample 10-13sec/state
- 1027 years to sample
Two‐state thermodynamics
To all appearances, the system (often) behaves as if there are only two thermodynamic states.

Entropy/Enthalpy
ΔG is a delicate balance of two large opposing energy contributions ΔH and TΔS.

| Reprinted with permission from N. T. Southall, K. A. Dill and A. D. J. Haymet, J. Phys. Chem. B 106, 521-533 (2002). Copyright 2002 American Chemical Society. | Reprinted from James Chou (2008). http://cmcd.hms.harvard.edu/activiti...1/lecture7.pdf. |
Cooperativity underlies these observations
Probability of forming one contact is higher if another contact is formed.
- Zipping
- Hydrophobic collapse

Reprinted from K. A. Dill, K. M. Fiebig and H. S. Chan, Proc. Natl. Acad. Sci. U. S. A. 90,1942-1946 (1993). Copyright 1993 PNAS.
Protein Folding Conceptual Pictures
Traditional pictures rooted in classical thermodynamics and reaction kinetics.
- Postulate particular sequence of events.
- Focus on importance of a certain physical effect.
- Framework or kinetic zipper
- Hydrophobic collapse
- Nucleation–condensation
Framework/Kinetic Zipper Model
- Observation from peptides: secondary structures fold rapidly following nucleation.
- Secondary structure formation precedes tertiary organization.
- Emphasis:
- Hierarchy and pathway
- Focus on backbone, secondary structure
Hydrophobic Collapse
- Observation: protein structure has hydrophobic residues buried in center and hydrophilic groups near surface.
- An extended chain rapidly collapses to bury hydrophobic groups and thereby speeds search for native contacts.
- Collapsed state: molten globule
- Secondary and tertiary structure form together following collapse.

Nucleation–Condensation
Nucleation of tertiary native contacts is important first step, and structure condenses around that.
Some observations so far:
- Importance of collective coordinates
- Big challenge: We don’t know much about the unfolded state.
______________________________________________________
- C. Levinthal, Are there pathways for protein folding?, J. Chim. Phys. Phys.-Chim. Biol. 65, 44-45 (1968).
- 20.1: Models for Simulating Folding
- Our study of folding mechanism and the statistical mechanical relationship between structure and stability have been guided by models. Of these, simple reductionist models guided the conceptual development from the statistical mechanics side, since full atom simulations were initially intractable. We will focus on the simple models.


