15.2: Facilitated Diffusion
- Page ID
- 294339
Facilitated diffusion is a type of dimensionality reduction that has been used to describe the motion of transcription factors and regulatory proteins looking for their binding target on DNA.1
E.coli Lac Repressor
Experiments by Riggs et al. showed that E. coli Lac repressor finds its binding site about one hundred times faster than expected by 3D diffusion.2 They measured ka=7×109 M−1 s−1, which is 100–1000 times faster than typical rates. The calculated diffusion-limited association rate from the Smoluchowski equation is ka≈108 M−1 s−1 using estimated values of D≈5×10−7 cm2 s−1 and R≈5×10−8 cm. Berg and von Hippel theoretically described the possible ways in which nonspecific binding to DNA enabled more efficient one-dimensional motion coupled to three-dimensional transport.3
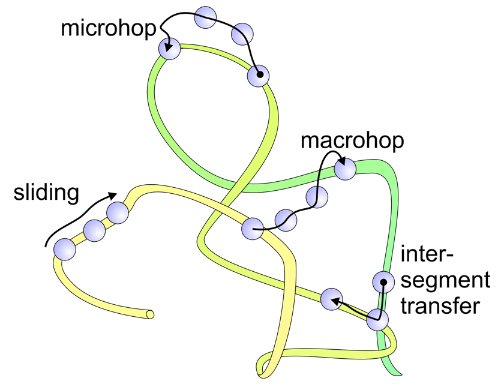
Many Possibilities for Locating Targets Diffusively: Coupled 1D+3D Diffusion
- Sliding (1D diffusion along chain as a result of nonspecific interaction)
- Microhop (local translocation with free diffusion)
- Macrohop (...to distal segment via free diffusion)
- Intersegmental transfer at crossing—varies with DNA dynamics
Consider Coupled Sliding and Diffusion: The Steady‐State Solution
The transcription factor diffuses in 1D along DNA with the objective of locating a specific binding site. The association of the protein and DNA at all points is governed by a nonspecific interaction. Sliding requires a balance of nonspecific attractive forces that are not too strong (or the protein will not move) or too weak (or it will not stay bound). The nonspecific interaction is governed by an equilibrium constant and exchange rates between the bound and free forms:
\[ F \overset{k_a}{\underset{k_d}{\rightleftharpoons}} B \qquad \qquad \qquad K = \dfrac{k_a}{k_d} = \dfrac{\overline{\tau}_{1D}}{\overline{\tau}_{3D}} \nonumber \]
We can also think of this equilibrium constant in terms of the average times spent diffusing in 1D or 3D. The protein stays bound for a period of time dictated by the dissociation rate kd. It can then diffuse in 3D until reaching a contact with DNA again, at a point which may be short range in distance but widely separated in sequence.
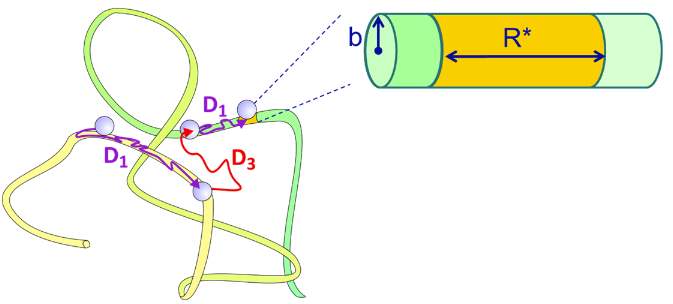
The target for the transcription factor search can be much larger that the physical size of the binding sequence. Since the 1D sliding is the efficient route to finding the binding site, the target size is effectively covered by the mean 1D diffusion length of the protein, that is, the average distance over which the protein will diffuse in 1D before it dissociates. Since one can express the average time that a protein remains bound as \(\overline{\tau}_{1D}=k^{-1}_d\), the target will have DNA contour length of
\[ R^*=\left( \dfrac{4D_1}{k_d} \right)^{1/2} \nonumber \]
If the DNA is treated as an infinitely long cylinder with radius b, and the protein is considered to have a uniform probability of nonspecifically associating with the entire surface of the DNA, then one can solve for the steady-state solution for the diffusion equation, assuming a completely absorbing target. The rate constant for specific binding to the target has been determined as
\[ \eta = \dfrac{D_1K'}{D_3b} \nonumber \]
where K' is the equilibrium constant for nonspecific binding per unit surface area of the cylinder (M–1 cm–2 or cm). We can express the equilibrium constant per base-pair as \(K=2\pi \ell bK'\), where \(\ell \) is the length of a base pair along the contour of the DNA. The association rate will be given by the product of kbind and the concentration of protein.
_________________________________
- P. H. von Hippel and O. G. Berg, Facilitated target location in biological systems, J. Biol. Chem. 264 (2), 675–678 (1989).
- A. D. Riggs, S. Bourgeois and M. Cohn, The lac represser-operator interaction, J. Mol. Biol. 53 (3), 401–417 (1970); Y. M. Wang, R. H. Austin and E. C. Cox, Single molecule measurements of repressor protein 1D diffusion on DNA, Phys. Rev. Lett. 97 (4), 048302 (2006).
- O. G. Berg, R. B. Winter and P. H. Von Hippel, Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory, Biochemistry 20 (24), 6929–6948 (1981).


