11.8: Supramolecular assemblies
- Page ID
- 139305
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Supramolecular assemblies are a different type of large structure, related to macromolecules. In a supermolecular assembly, parts of the structure are held together by very strong interactions, but not necessarily by covalent bonds. For example, a polymer in which chains are held together by physical crosslinks such as hydrogen bonds may be considered to form a supermolecular assembly.
DNA is an example of a supramolecular assembly. Each strand of DNA is an alternating sugar-phosphate copolymer. Along this backbone, there are pendant "base" units, adenine, cytosine, guanine or thymine.
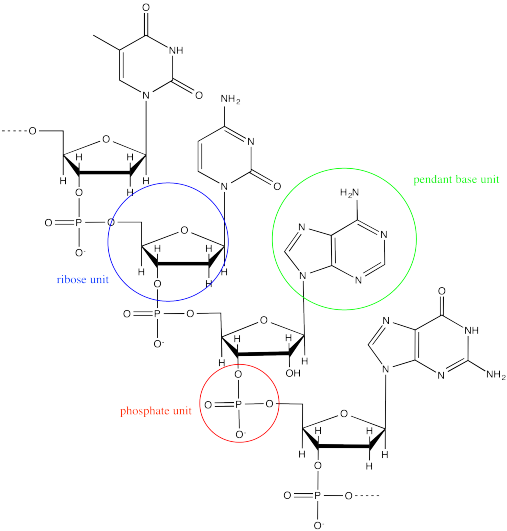
Two DNA strands form a helical supramolecular assembly through hydrogen bonding interactions that form between the bases. Thymine can hydrogen bond preferentially with adenine.
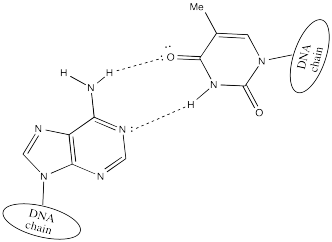
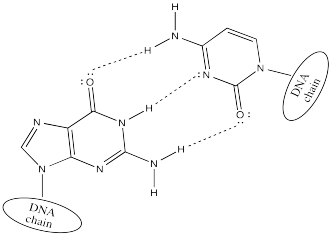
Cytosine hydrogen bonds preferentially with guanine.
There are a number of different ways in which synthetic supramolecular assemblies can be formed. These kinds of structures have been getting increased attention in recent years as researchers try to develop new materials with useful properties. There are a few common methods through which the assemblies are formed. We will look at hydrogen bonding, host-guest chemistry and coordination chemistry.
Hydrogen Bonding
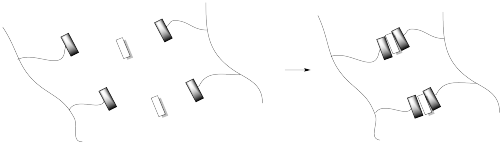
Hydrogen bonding is a common way to build supramoleucular assemblies. Bert Meijer at Eindhoven Technical University in the Netherlands has developed a number of systems that use this approach. For example, graft copolymers have been assembled in his lab by hanging pendant chains onto a polymer backbone via hydrogen bonds.
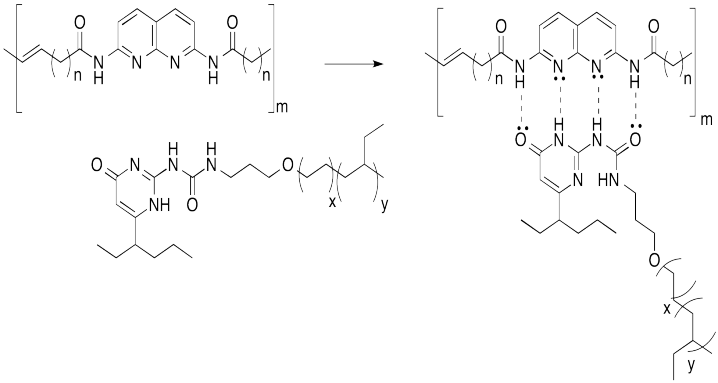
This approach can be pictured in cartoon, below, in which the hydrogen bonding elements are boxes and the lines are flexible chains or tails.
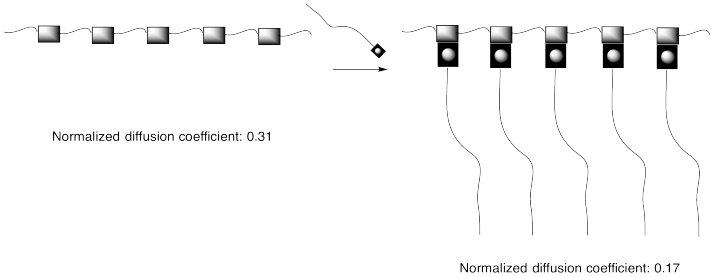
Problem MM8.1.
The diffusion coefficients cited in the picture above are related to how easily compounds can move through a solution. The higher the diffusion coefficient, the more easily the compound moves through solution.
a) How is the diffusion coefficient related to viscosity?
b) Explain the difference between the diffusion coefficientsof the two compounds above.
Host-Guest Complexes
Another approach to supramolecular assemblies is to exploit host-guest interactions to bring different parts of the assembly together. Host-guest complexes act a little bit like enzymes and their substrates in biology, or like materials bound inside zeolites. The idea is that one molecule has a cavity or opening big enough for another molecule to fit inside. It's a little like a basketball fitting into a peach basket.
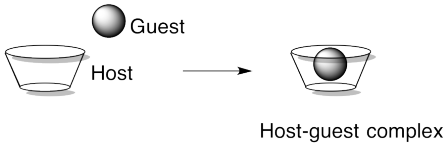
There might be a physical attraction (an intermolecular force) between a host and a guest, but there doesn't have to be. Julius Rebek at the Scripps Institute has developed host complexes that will hold normally aloof guests such as N2. Part of the reason guests can become comfortably ensconced without intermolecular attractions involves the fact that the molecule inside would have to take a very particular path in order to escape.
A number of host compounds are shown below. Each of them is designed to have an enclosed space of some kind. In some cases it's no more than a ring, but that is enough to keep the guest from escaping on those sides.
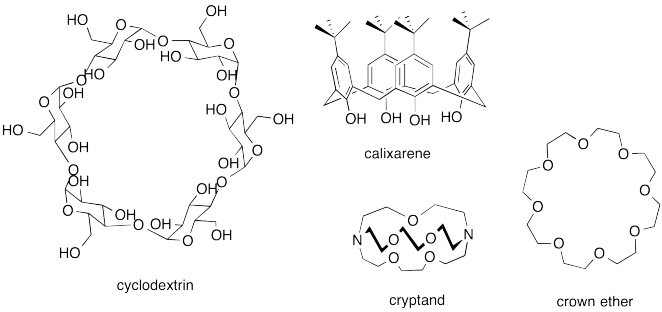
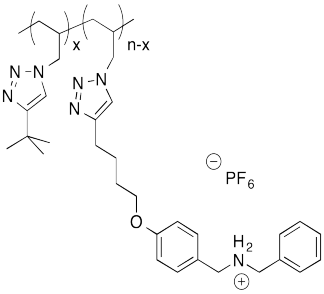
Problem MM8.2.
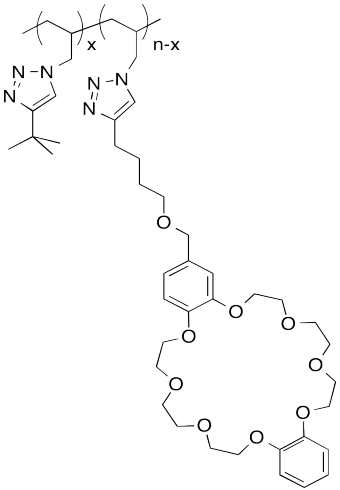
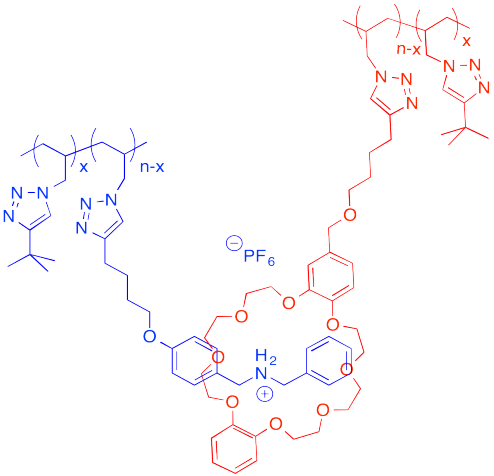
The following host-guest system was designed in the lab of Taichi Ikeda at the National Institute for Materials Science in Japan. In the polymer notation, "x" and "n-x" are just numbers of repeating units. When solutions of the two polymers are mixed together, an elastic gel is formed.
a) Draw a cartoon of each of the polymers and of the host-guest linked assembly.
b) What makes this host and guest especially attractive for each other?
c) Describe the structure of this assembly, in terms of polymer architectures seen previously.
d) Why is the gel elastic?
e) This gel is "self-healing": it can be cut with a knife, but the pieces can then be stuck back together again. Explain how this re-attachment occurs.
Coordination Compounds
If you don't know about coordination compounds, you might want to start with Lewis acid-base complexes.
A number of such assemblies have been reported from the lab of Stephen Craig at Duke University. For example, mixing a solution of certain palladium compounds into a solution of poly(vinylpyridine) (PVP) produces a thickening of the liquid into a honey-like consistency.
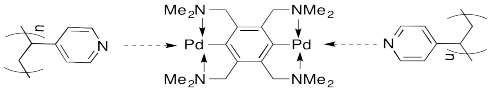
As a cartoon, this system can be represented as shown below. The palladium complexes form physical crosslinks between the pendant pyridine units on the polymer chains.
Problem MM8.3.
This is a challenge problem for students of Chem 251.
Craig's lab reported the following kinetic data for the poly(vinylpyridine) / MLn system in DMSO (dimethylsulfoxide) solvent. Remember, MLn just means "coordination complex".
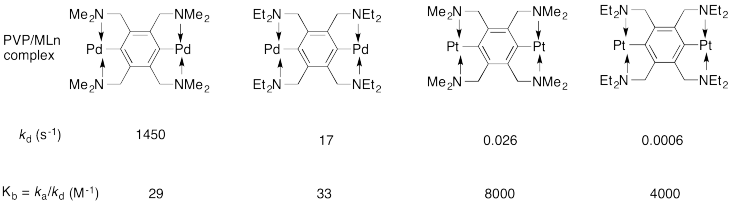
a) The rate constant kd refers to the dissociation of a PVP/MLn linkage. Show a likely mechanism for that reaction.
b) Explain why the reaction is faster with the methyl-substituted ligand than the ethyl-substituted one.
c) Is Kb the equilibrium constant for formation of the PVP/MLn linkage or for dissociation of the linkage?
d) Explain the large difference in values of Kb between the palladium complexes and the platinum complexes.


