Immunoassays
- Page ID
- 296399
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction to Immunoassays
For the questions below, you have the following pieces available to build an immunoassay. You may use some or all of them, in any combination, and may use more than one instance of each piece if needed. You can also wash with buffer in between steps to remove excess of the reagent that you just added.
- You are trying to detect protein analyte A in a complex biological sample that contains many other analytes, using a heterogeneous immunoassay. Heterogeneous means that the assay is conducted on a solid surface, e.g. a plastic plate. You will use an enzymatic reporter.
- What type of immobilization strategy is preferable? Direct OR Indirect Capture
Why?
- Sketch complete schematics for two versions of this assay.
Direct detection Indirect detection
- What is another name for this type of immunoassay?
- What must be true about the capture and detection antibodies in this assay, with respect to their binding sites on the antigen, to have a successful assay?
- Which must be present in molar excess, the antibodies or the antigen, to make the signal linear with the concentration of analyte?
- What type of immobilization strategy is preferable? Direct OR Indirect Capture
- You recently produced the analyte A recombinantly, and need to quantify its concentration in solution. You will use a heterogeneous immunoassay to do so.
- For the simplest possible assay, what should you adsorb to the plate?
- Sketch complete schematics for two versions of this assay, using direct detection:
Enzymatic reporter Conjugated fluorophore as a reporter
- For the simplest possible assay, what should you adsorb to the plate?
- Comparing immunoassay strategies. For each of the options below, describe the advantages/disadvantages of each, or when each option might be most suitable.
- Direct capture vs indirect capture of the antigen.
- Direct vs indirect detection.
- Linear (e.g. one or more conjugated fluorophores) vs enzymatic reporter.
- Direct capture vs indirect capture of the antigen.
- Sketch a plot of what typical data from a non-competitive immunoassay looks like. Label the axes. Demarcate the linear range of the assay as well as what happens above and below it.
Antibody immobilization and labelling strategies
Learning Goals
Students will be able to:
- Describe the differences between oriented and non-oriented immobilization strategies and recognize the impact of orientation of protein function.
- Describe the mechanism of non-covalent adhesion of a biomolecule to a surface, and define experimental parameters that affect the efficiency of adhesion.
- Describe multiple options for oriented antibody binding using biomolecules, and recognize their limitations.
- Describe biotin-streptavidin binding and its uses for protein immobilization.
- Look up and find chemical and biochemical information using online resources.
Part I. What do we mean random vs oriented attachment?
- Draw schematics of antibodies attached in a non-oriented (random) or oriented manner on a surface.
______________________ _______________________
Non-oriented Oriented
Part II. Strategies for non-oriented attachment to a surface:
- Non-covalent interactions
- There are two common methods, both of which add protein in buffer to the surface and incubate. Incubation can be either:
- Overnight at 4 °C, or
- 2 hours at 37 °C
For many proteins, the efficiency of surface immobilization is similar by these two methods. Why might incubating at higher temperature enable a shorter time?
- Although many proteins are efficiently adsorbed to a plastic plate at pH 7.4, some are better adsorbed at high pH (e.g. pH 9.6). Why? Consider the biochemistry of a protein molecule.
- There are two common methods, both of which add protein in buffer to the surface and incubate. Incubation can be either:
- Covalent attachment and high-affinity interactions
Previously you learned about methods to covalently add a modification to a protein (bioconjugation chemistry). Here we will consider an alternative, non-covalent but essentially irreversible strategy for antibody immobilization: the biotin-streptavidin linkage.
A measure of affinity that you will learn about later in the course is KD, the dissociation constant. A smaller KD indicates a higher affinity interaction. Most antibodies have KD values in the low micromolar (10-6 M) to nanomolar (10-7 M to 10-9 M) range. For antibodies, a micromolar KD (10-6 M) is considered low affinity, low nanomolar KD (10-9 M) is considered high affinity, and picomolar KD (10-12 M) is exceptionally high affinity.
-
What does the KD of ~1x10-14 M for biotin and streptavidin tell you about their interaction?
- Imagine you conjugate biotin to an antibody via the NHS chemistry you learned previously (using a biotin-NHS reagent). Do you have control over where the biotin molecules attach to the antibody? Why or why not?
- Consider how the interaction between avidin and an antibody that was biotinylated using a biotin-NHS reagent might be leveraged to help with antibody immobilization on a plate. Draw and label a diagram with the following components: antibody, plate, biotin, avidin. Considering your response to (b), make clear in your schematic why the attachment is considered “non-oriented”.
-
- Brainstorm four mechanisms by which non-oriented covalent binding to a surface might decrease binding activity of the antibody. It may be useful to sketch the structure of the antibody bound to the surface.
Part III. Strategies for oriented attachment:
- Researchers frequently take advantage of biological proteins and antibodies that bind the Fc region. Go to ThermoFisher’s “IgG-binding proteins” page and the links therein to learn more.
-
What is the origin of Proteins A and G?
- Do all IgG antibodies necessarily bind to protein A with the same affinity as to Protein G? Comment.
- Your labmates use Protein G successfully to capture their antibody of interest, which is a human IgG. You are working with a rat IgG. Is it safe to assume that your antibody will also be bound by protein G? Support your prediction by looking up the binding strength in each case.
-
- Biotinylation of an antibody can also be performed in a site-specific manner, by oxidizing the carbohydrates at the glycosylation sites to an aldehyde, then reacting with a nucleophilic hydrazide-biotin reagent. (more info at Pierce Protein Methods handbook).
Why would this strategy for biotinylation be more likely to produce oriented attachment then biotinylation by NHS chemistry? It may be useful to draw a sketch of the two scenarios.
- Consider the following data, from Peluso et al (Anal Biochem, 2003). An antibody (left side of figure) or its Fab fragment (right) was bound either randomly (top) or in an oriented manner (bottom) on the surface of an ELISA plate. Then (figure below), a saturating concentration of the target antigen was incubated on the plate, to quantify the total analyte-binding capacity of the system (A). The density of available binding sites (B) was quantified by surface plasmon resonance. The ratio of analyte binding to binding site density was calculated and called specific activity (C).

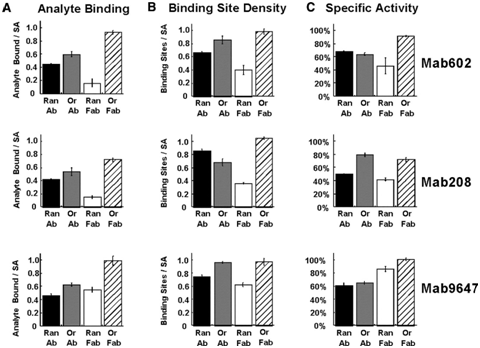
Caption: Effects of various linkage strategies on binding site densities and binding activities of three different monoclonal antibodies (Mab). Specific activity is defined as the density of bound analyte (A) divided by the binding site density (B). To saturate the active sites, 100 nM analyte concentrations were employed for Mab208 and Mab602 while 1 μM analyte concentration was used for Mab9647. -
Examine this data and draw conclusions about the effect of oriented vs random binding on whole antibodies.
- Examine this data and draw conclusions about the effect of oriented vs random binding on Fab fragments.
- Now compare the data for whole antibodies versus Fab fragments. Why might orientation have a larger impact on binding for Fab fragments than for whole antibodies?
-
Immunoassay readouts – In-class activity
Learning Goals
Students will be able to:
- Recognize the structures and properties of common colorimetric ELISA substrates.
- Find product information online on vendor websites.
- Describe alternative methods of detection for immunoassays and discuss advantages and disadvantages of each method.
After carefully choosing antibodies to detect an analyte, how do you best “read out” the assay?
- ELISA – enzymatic readout
Search online to find the following information for four common colorimetric ELISA substrates. Good resources may include vendor websites, chemistry sites, or published reviews or papers.
Full chemical name
and
StructureEnzyme
Color of product
Max Abs λ (nm)
Kinetics: "fast" or "slow"?
TMB
ABTS
PNPP
Primary reference(s) used:
- Non-enzymatic readouts
The most common non-enzymatic readout is fluorescence, using fluorescent dyes conjugated to the detection antibody or to streptavidin. However, many other creative options have been developed, for example for a readout that is visible by eye.
Search online to research one of the following methods in terms of how it has been used as a readout for an immunoassay: Nanoparticle aggregation, turbidity, silver precipitation. (circle)
Choose one immunoassay design that you find interesting. Draw a schematic of the immunoassay complex and depict how the detection method works. Include a 1-sentence caption that describes what they are detecting and the premise of the assay.
What are the advantages of this method compared to ELISA or fluorescent labels?
What are the disadvantages of this method compared to ELISA or fluorescent labels?
Primary reference(s) used:
Contributors and Attributions
- Rebecca Pompano, University of Virginia (rrp2z@virginia.edu)
- Sourced from the Analytical Sciences Digital Library


