2.7: Day 15- Condensation Polymers, Proteins
- Page ID
- 371687
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)14
Day 15: Condensation Polymers, Proteins
D15.1 Condensation Polymers
A condensation polymer is a polymer formed via a condensation reaction (see Section D13.5). The very small molecule produced in each condensation can be H2O, HCl, or some other simple molecule. Condensation polymerizations often, but not always, combines two different monomers in an alternating structure.
Condensation polymers usually grow by forming ester or amide linkages, where new C-O or C-N σ bonds form to link monomers. A typical monomer has a functional group at each end of the molecule. When one end of the monomer reacts and is added onto a polymer chain, the functional group at the other end remains and allows for further reaction to lengthen the polymer chain.
D15.2 Polyesters
A polyester is a polymer where the individual units are held together by ester linkages. For example, the common polyester polyethylene terephthalate (abbreviated as PET or PETE; has brand names such as Dacron or Mylar) consists of polymer chains formed from terephthalic acid (benzene-1,4-dicarboxylic acid) and ethylene glycol (ethane-1,2-diol) monomers.
The condensation reaction that results in the formation of the ester linkage in PET is shown in Figure 1. Repeated condensation reactions result in the step-wise growth of a polymer chain.
The repeating unit of PET is

The presence of ester groups, a polar functional group, on the polymer increases the intermolecular forces between polymer chains, and thereby increases the crystallinity and tensile strength of the polymer. PET makes for excellent fibers and are used in many fabrics. It is also used to make bottles for soda and other beverages. Polyester is biologically inert, so a knitted polyester tube can be used in surgery to repair or replace diseased sections of blood vessels.
D15.3 Polyamides
A polyamide is a polymer in which the individual units are held together by amide linkages. For example, nylon 66 is obtained from the monomers 1,6-hexanediamine and hexanedioic acid. The condensation reaction between these two monomers forms an amide linkage (highlighted):
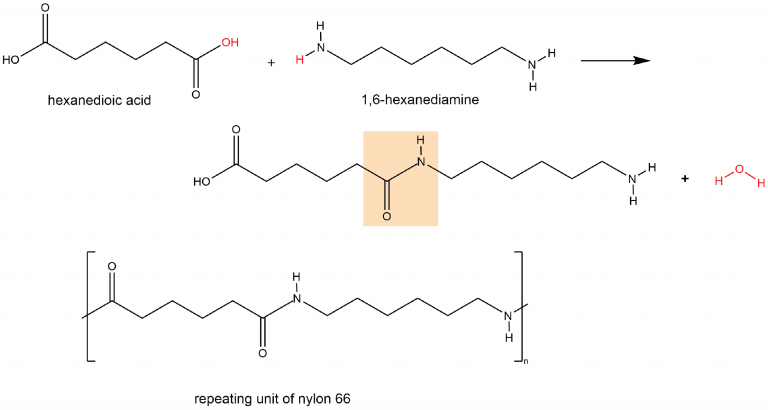
On one end, the product molecule has a carboxylic acid group, which can undergo a condensation reaction with another 1,6-hexanediamine molecule. On the other end there is an amine group, which can react with another hexanedioic acid molecule. Such continuous condensation reactions lead to the formation of a nylon 66 polymer strand, the repeating unit of which is also shown above.
The “66” in nylon 66 stands for the six carbon atoms in each of the monomer molecules. Other nylons have different numbers of carbon atoms in the monomer molecules, such as nylon 510, which has 5 carbon atoms in the diamine (1,5-pentanediamine) and 10 carbon atoms in the diacid (decanedioic acid).
Nylon makes extremely strong threads and fibers because in addition to London dispersion forces and dipole-dipole attractions, there are hydrogen bonds between the polymer chains. Specifically, a hydrogen bond can form between a N-H in one strand and a carbonyl O lone pair in a neighboring strand.

If you pull on both ends of a nylon thread, after a slight stretch, it will resist breaking because of the strong IMFs that hold neighboring chains together.
Kevlar is a synthetic polymer made from 1,4-phenylenediamine and terephthaloyl chloride monomers. The byproduct of this condensation reaction is HCl.

The material has a high tensile-strength to weight ratio (it is about 5 times stronger than an equal weight of steel), making it useful for many applications from bicycle tires to sails to body armor.
Similar to nylon, part of Kevlar’s strength is due to dipole-dipole interactions and hydrogen bonding (Figure 3) that increase intermolecular forces between polymer strands.

Notice that compared to nylon 66, the density of hydrogen bonds in Kevlar is higher, imparting stronger intermolecular forces.
Furthermore, Kevlar also has stronger London dispersion forces. Consider the repeating unit of Kevlar highlighted in Figure 3. It is a planar structure, with planar benzene rings connected by planar amide groups. Hence a single layer of Kevlar, as shown in Figure 3, is planar. When layers of Kevlar are placed on top of each other, they can be stacked very efficiently, allowing for maximal London dispersion forces.
In addition to its better-known uses, Kevlar is used in cryogenic applications because of its very low thermal conductivity. Kevlar maintains its high strength when cooled to liquid nitrogen temperatures (–196 °C). Many other plastics become brittle and break at that temperature.
D15.4 Proteins
Proteins are condensation polymers made from amino acids. An amino acid consists of a carbon atom (called the α carbon, shown in red below) bonded to a hydrogen atom, an amine group, a carboxylic acid group, and an R group, often called the side chain .

Notice that the α carbon is chiral when the “R” group differs from the other three groups. About 500 naturally occurring amino acids are known, although only twenty are found in protein molecules in humans. Of these twenty amino acids, nineteen have a chiral α carbon.
Amino acids are linked together in proteins by amide linkages formed via condensation reactions. An amide linkage in a protein is often called a peptide bond.

Note that the reaction does not involve the α carbon atom, and the chirality at the α carbon is the same after the reaction as it was before. (In the equation above, R2 is depicted with a dashed bond because the amino acid shown in blue has been flipped—rotated 180° around a horizontal axis—from the reactant to the product.)
Like all amide functional groups, the amide linkages in a protein strand are planar. The α carbon atoms, however, have tetrahedral local geometry. This molecular chain of α carbons connected by amide groups, is referred to as the protein backbone. It is the main chain of a strand of protein. The amino acid R groups are called side chains because they are attached to, but branch off from, the main chain.
15.5 Amino Acids
The twenty amino acids are often sorted into four groups—hydrophobic, polar but uncharged, basic, and acidic—depending on the nature of their side chains. The nine amino acids that have hydrophobic side chains are shown in Figure 4:

These hydrophobic side chains are composed mostly of carbon and hydrogen, have very small dipole moments, and tend not to mix with water. As we will see, this characteristic has important implications for a protein’s structure.
The six amino acids shown in Figure 5 have side chains that are polar but neither acidic nor basic.

The three amino acids in Figure 6 have side chains containing functional groups that are basic at neutral pH:

Activity 1: What Part of the Histidine Side Chain Is Most Basic?
And finally, Figure 7 shows two amino acids that have acidic side chains containing the carboxylic acid functional group:

D15.6 Protein Structure
Protein molecules are complicated: there are 20 different kinds of monomers that can be linked to form a huge number of different polymer chains. Thus protein structures are classified in several ways. Fundamental is the order in which the monomers are linked: the primary structure. Long protein backbones can adopt unique three-dimensional secondary and tertiary structures, both of which are dependent on the primary structure. When two or more protein strands of proteins are held together by intermolecular forces, they can also adopt specific quaternary structures.
Primary Structure
A protein molecule is a polyamide (polypeptide), but because there are 20 different amino-acid monomers, a protein is a copolymer on steroids. Although the repeating unit is the amino-acid structure, there are 20 possibilities for each side chain (highlighted in red below):

Thus a large number of different protein-molecule structures are possible. Each possible copolymer structure has a unique sequence of side chains. This specific sequence of amino acids is called the protein’s primary structure.
By convention, protein sequences are written from the end with the free amine group (the N terminus) to the end with the free carboxyl group (the C terminus). For example, Ala-Ser (alanine-serine) differs from Ser-Ala (serine-alanine).

Secondary Structure
A protein’s secondary structure is the three dimensional shape of small segments of proteins. In secondary structures amino acids that are not next to each other along the backbone are close enough to each other for hydrogen bonding. The type of secondary structure is defined by the pattern of hydrogen bonds between amide groups that are not adjacent in the protein’s backbone. Two common elements of secondary structure, which are present in over 60% of known proteins, are the α-helix and β-sheet.
The α-helix has a helical structure where there are, on average, 3.6 amino acids per turn of the helix. All the side chains project out from the helical axis.

An α-helix has a regular pattern of hydrogen bonding between backbone amide groups, where the nth amino acid is hydrogen-bonded to the (n+4)th amino acid. For example, as shown in Figure 9, the C=O of amino acid 1 forms a hydrogen bond with N-H of amino acid 5; the C=O of amino acid 2 forms a hydrogen bond with N-H of amino acid 6; etc. This regularly repeating hydrogen bonding is a prominent characteristic of an α-helix.

The β-sheet or β-pleated sheet consists of stretches of the protein chain connected side-by-side by hydrogen bonds between backbone amide groups.

Such backbone hydrogen bonding gives rise to a general pleated sheet appearance, hence the name. The side chains of the amino acids alternate above and below the β-sheet (Figure 11).

The numerous backbone hydrogen bonds stabilize both the α helix and β sheet structures. These structures can combine to form various larger structures, called motifs, that are found in more than one protein structure.
Tertiary Structure
The tertiary structure of a protein is the overall three-dimensional shape of the protein. It is how α-helices, β-sheets, and motifs come together to form the overall structure. Tertiary structures are often defined by various intermolecular interactions involving the protein’s side-chains as well as the environment the protein is in.
Two general kinds of proteins are found in cells, water soluble and water insoluble proteins. Water soluble proteins, which include enzymes and transport proteins, are found free in cellular compartments such as the cytoplasm, nucleus, or endoplasmic reticulum.
In the tertiary structure of water-soluble proteins, most hydrophobic side-chain groups are in the interior of the protein and away from water, while most polar side-chain groups are mostly kept on the exterior and therefore exposed to water. (The backbone amide groups, while polar and capable of forming hydrogen bonds, do not interact much with water because they are already engaged in hydrogen bonding with each other in α helices and β sheets.)

The water-insoluble proteins include membrane channels, pumps, and receptors. These proteins are found in lipid bilayers. A description of how and why these proteins are associated with membranes is in Section D16.7.
One important determinant of tertiary structure in some proteins is the disulfide bond. It is similar to the cross-linking we’ve discussed for addition polymers. A protein can form disulfide bonds when cysteines in different parts of its strand are linked by a covalent bond between the sulfur atoms in their side-chains (Figure 13).
 Figure 13. Disulfide bonds. Left: A disulfide bond (red) connects two cysteine side chains along the protein backbone. Right: Three disulfide bonds are shown in yellow. The backbone is shown as magenta ribbons and cysteines are shown as ball-and-stick models. The six cysteines in this protein form three S-S linkages. (PDB: 6RVA)
Figure 13. Disulfide bonds. Left: A disulfide bond (red) connects two cysteine side chains along the protein backbone. Right: Three disulfide bonds are shown in yellow. The backbone is shown as magenta ribbons and cysteines are shown as ball-and-stick models. The six cysteines in this protein form three S-S linkages. (PDB: 6RVA)
Quaternary Structure
Quaternary structure arises when more than one protein chain is present and the chains form a larger structure. For example, the hemoglobin in your blood is composed of four chains (colored magenta, blue, aqua, and tan below) which form two subunits.
When oxygen binds to one subunit of hemoglobin, the interactions between the subunits change. In other words, oxygen binding affects the quaternary structure. Quaternary structure is the usual mechanism for the processes known as allostery (in which binding of a molecule at one site on a protein has effects at other sites far away from that site) and cooperativity (in which binding of a molecule at one site in a multi-subunit protein increases or decreases the likelihood that another molecule will bind at another site of the protein).
D15.7 Protein Folding and Denaturation
How do proteins fold into the complicated three-dimensional structures? This is a very active area of research in chemistry and biochemistry right now. The first hint came from the work of Christian Anfinsen on the protein ribonuclease, which breaks down RNA molecules. Anfinsen discovered that after treating ribonuclease with high concentrations of certain chemicals that cause proteins to unfold and lose their tertiary and secondary structure, the ribonuclease no longer broke down RNA. Moreover, if the chemicals were removed, the ribonuclease would spontaneously recover nearly all its RNA-hydrolyzing activity, without needing any other cellular components. Anfinsen concluded that the primary structure of a protein completely determines its three-dimensional structure at the secondary, tertiary, and quaternary levels.
Scientists are still trying to learn how the primary structure of a protein determines its other levels of structure. They have determined the primary forces that stabilize a protein’s three-dimensional structure are:
- Sequestration of hydrophobic side-chains away from water (for example, in the interior of water-soluble proteins)
- Maximizing London dispersion interactions (minimizing open spaces) in the interior of proteins
- Maximizing hydrogen bonding (for example, in α-helices or β-sheets)
- Attractions between negatively and positively charged sites formed when acidic and basic side-chains lose and gain H+ ions
The process that Anfinsen used is called denaturation, in which proteins lose the quaternary, tertiary, and secondary structures which is present in their native state. Proteins can be denatured by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent (e.g., alcohol or chloroform), radiation, or heat. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups.
Protein folding is key to whether a protein can do its job correctly; it must be folded into the right shape to function. However, hydrogen bonds, which play a big part in folding, are weak compared to covalent bonds and thus can easily be affected by heat, acidity, varying salt concentrations, and other stresses. This is one reason why homeostasis is physiologically necessary in many life forms.
https://mediaspace.wisc.edu/id/0_o4v8ja01?playerId=25717641
Day 15 Pre-class Podia Problem: Nylon
The repeating unit of nylon-6,10 has this structure.

Answer these questions:
- Write the structures of two monomers that could be combined to form nylon-6,10.
- List all the intermolecular forces that exist between polymer chains in nylon-6,10.
- Depending on the orientation of adjacent polymer chains, intermolecular forces between nylon-6,10 chains might be stronger or weaker. Draw structures of two nylon-6,10 repeating units, one above and one below the structure given. Orient the structures you draw to maximize intermolecular forces.
- Indicate in your drawing which types of intermolecular forces are important between which parts of the molecules. Also, write an explanation in words describing which forces are important where and why.
Two days before the next whole-class session, this Podia question will become live on Podia, where you can submit your answer.
Comments. If you found any inconsistencies, errors, or other things you would like to report about this module, please use this link to report them. A similar link will be included in each day’s material. We appreciate your comments.


