IB4. Carbohydrates
- Page ID
- 4058
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)IB4. Intermolecular Forces in Biology: Carbohydrates (contributed by Henry Jakubowski)
Carbohydrates (CHOs) are among the most complex of biological molecules. If you have ever "counted" your carbs, you know that one biological function of CHOs is to store and, on oxidation, provide energy to the body for required functions. Instead of concentrating on how CHOs are used for energy production, we will focus predominantly on their structures, which allows them to elicit their main function, which is to provide binding interactions with other biomolecules, either in solution or on cell membranes. Binding, promoted through IMFs, initiate and modulate all biological interactions and processes. First, we will consider the structure of simple and more complex carbohydrates.
Monosaccharides
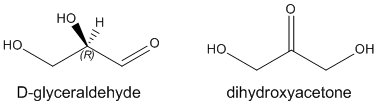
The simplest carbohydrates are monosaccharides, small organic molecules that contain more than one OH (alcohol) group and a single aldehyde (RCOH) or ketone (RCOR). Hence the simplest monosaccharides, glyceraldehyde and dihydroxyacetone, contain three carbons. In contrast to the alkane hydrocarbons which have the generic formula CnH2n+2, simple monosaccharides have the generic formula Cn(H2O)n which corresponds to their designation as carbo(hydrates).
Glyceraldehyde has one stereocenter. The naturally occurring isomer shown above is R glyceraldehyde. Instead of using the R/S systems to denote the absolute configuration of specific stereocenters in sugars (as well as amino acids), biochemists often use the D/L system. The D designation on sugars refers to any monosaccharide whose last stereocenter (or in the case of glyceraldehyde the only stereocenter) has the same absolute configuration as r-glyceraldehyde. All naturally occurring amino acids have the L configuration as they have the same absolute configuration when compared to L-glyceraldehyde.
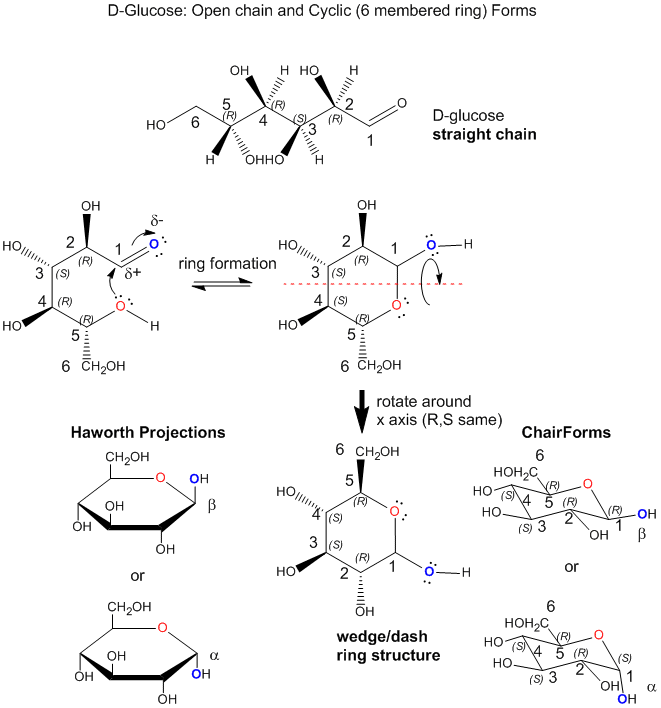
Monosaccharides can exist in solution in an equilibrium mixture of a straight chain and cyclic form. An example is shown below for the common sugar D-glucose (Glc). The R/S designation for each stereocenter is included to make it easier to see the orientation of each OH group as the straight chain form cyclizes. In the process of forming the 6-membered ring, a lone pair on the slightly negative OH on C5 forms a bond to the slightly positive C1, in a process that requires rotation around the various C-C to allow the OH on C5 to approach C1. Since the carbon can only have 4 bonds, bond formation to C1 from the OH on C5 causes the C=O (an aldehyde) to become C-OH (an alcohol). The chair, wedge/dash and "Haworth" projections of the resulting ring are shown. Note that the OH on C1 can point up (beta form, remembered by the phrase butterflies UP) or down (alpha form, remembered by ants DOWN). In the beta form, all ring substituents are in the equatorial position (in the chair form) and in alternating up and down forms in the wedge structure, making the beta form the most stable of all possible 6-membered cyclic forms of 6C sugars.
Since there are 4 stereocenters in a 6C sugar aldehyde, there are 24 or 16 possible stereoisomers. Most 6C sugars are D sugars which are defined to have the same R,S designation as D-glyceraldehyde at the last stereocenter in the sugar. Common 6C sugars are D-mannose (Man) and D-galactose (Gal). D-glucose, D-mannose, and D-galactose, all of which have the R configuration at C5, differ in the orientation of the OH groups in the ring structures at C2, C3, and C4. Hence they are diastereomers. The most common 6C sugar containing a ketone (RCOR) at C2 is fructose.
Since sugars are polyhydroxy aldehydes or ketones, can the OHs on C2, C3, and C4 also form a bond to C1 (as illustrated for the C5 OH above) and form rings. They can but the most common form is the 6-membered ring form shown above for D-glucose. How many atoms would be in the rings if the other OHs were involved in ring formation? Why is the 6-membered ring most abundant in nature?
Question
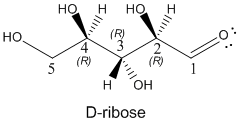
Draw the most stable form of the ring structure for the 5C sugar D-ribose, whose straight chain form is shown below. D-ribose is extremely important in biochemistry since its 5-membered ring form is found in RNA and a variant, deoxyribose, in which the OH on C2 is replaced with an H atom, is part of the main chain of DNA.
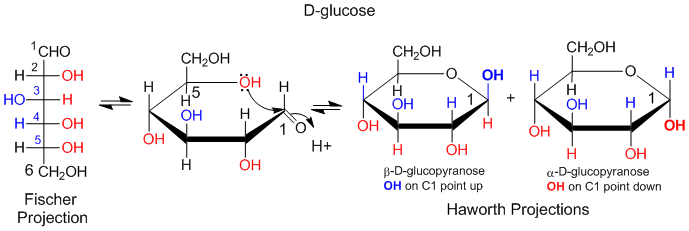
Another type of structural representation, the Fisher projection, is often used in texts. In this form, the structure is written in a vertical direction with vertical and horizontal lines (not wedges or dashes). It is simple to translate a Fisher project for simple sugars into a Haworth projection, and from there to a wedge/dash ring structure and then to a chair form. An example of how to convert a Fisher projection to a Haworth for D-glucose is shown below. If a OH group points to the right in the Fisher projection, it points down in the Haworth projections. If the OH points to the left, it points up in the Haworth projection.
Chemical Derivatives of Monosaccharides
Many derivatives of monosaccharides are found in nature. These include
- oxidized forms in which the aldehyde and/or alcohol functional groups are converted to carboxylic acids
- phosphorylated forms in which phosphate is added by ATP to form phosphoester derivatives
- amine derivatives such as glucosamine or galactosamine
- acetylated amine derivatives such as N-Acetyl-GlcNAc (GlcNAc) or GalNAc
- more complex condensation products of sugar derivatives with lactate (CH3CHOHCO2-) and pyruvate, (CH3COCO2- ) to form muramic acid and neuraminic acids, (also called sialic acids), respectively.
Di- and Polysaccharides
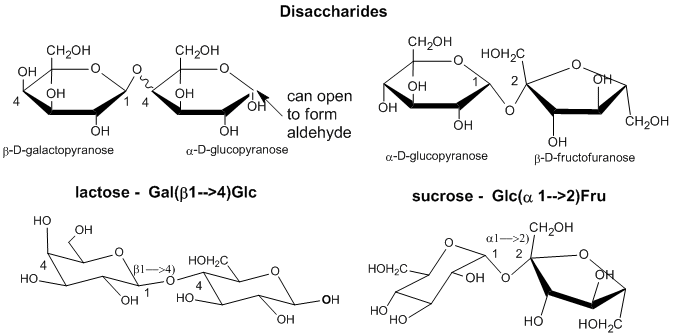
As with proteins, which are polymers of amino acid monomers, covalent bonds can form between different sugar monomers to form disaccharides, trisaccharides, and ultimately polysaccharides, as shown below for two disaccharides, lactose and sucrose..
Longer polysaccharides can be quite simple in structure, but given the diversity of monosaccharides with each having multiple OH groups that can be used to connect two monosaccharides in a polymer, the complexity of polysaccharide structure can be overwhelming to study and understand. Compare this to a protein, a polymer of amino acids each, with the exception of the side chains, have only two functional groups which are involved in the linkages between the amino acids.
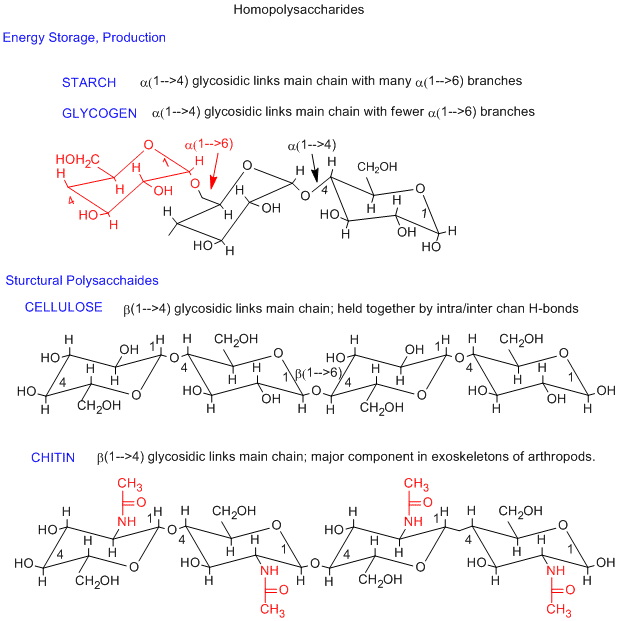
The simplest polysaccharides consist of only one monosaccharide repeating unit. Three examples of homopolysaccharides, glycogen/starch, cellulose, and chitin are shown below. The monomer in glycogen/starch and cellulose is glucose. The differences between them are in the linkage between the glucose monomers. In starch and glycogen, which are energy storage polysaccharides, the linkage is alpha 1-4 while in cellulose, the most abundant biomolecule, the linkage is beta 1-4. The beta linkage ensures that all bulky groups on the glucose chairs are in the more stable, equatorial position. This is yet another example of the fact that structure can account for properties and function of molecules. A small covalent change to glucose, the substitution of a CH3CONH- for a OH group on C2 of glucose gives the monosaccharide N-acetylglucosamine. This polymerizes, like cellulose, with a beta 1-4 linkage, to form a new polysaccharide, chitin, with markedly different properties than celluose. Chitin, is the main component of the exoskeleton of arthropods.
-
Jmol model of glycogen (similar to starch): Note that the the main chain twists (as indicated by the black chair forms in the figure above showing the main chain alpha 1-4 links), and thus forming a long helix. Triodiide, I3-, can fit into the opening of the helix, causing a solution of starch/glycogen to turn purple in this well known test for starch.
-
Jmol model of cellulose: Note the extended, non-helical structure of the polymer
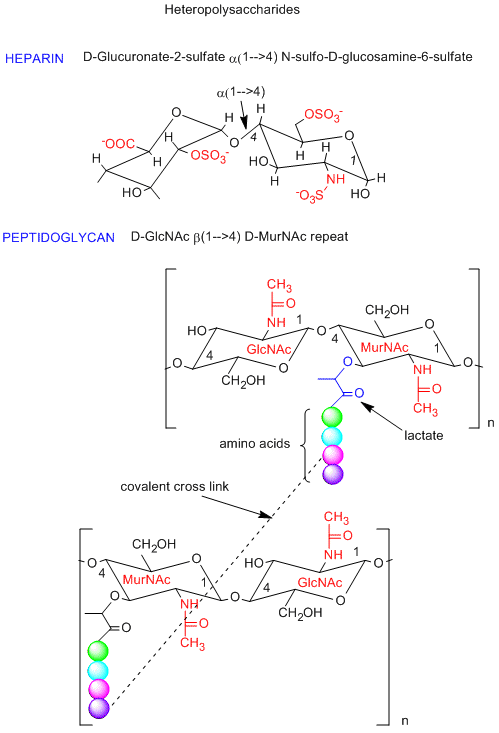
Polysaccharides can also be made of a repeating dissacharide unit, instead of a monosaccharide repeat in the homopolysaccharides shown above. One example is a glycosoaminoglycan called heparin (an anticoagulant, shown in the figure below) which is composed of a repeat of the dissacharide D-glucuronate-2-sulfate (alpha 1,4) GlcNSulfo-6-sulfate. This dissacharide consists of two glucoses that have been chemically modified. Note the charges on this repeat unit. Another example is the dissacharide repeat in the peptidoglycan (contains both CHO and amino acids) cell walls of bacteria (see figure below). The repeat again consists of chemically modified glucose units. Compare the repeat with the main component of the cytoskeleton, chitin. Nature has chosen variants of this repeat to create a rigid and protective exoskeleton and cell wall for arthropods and bacteria.
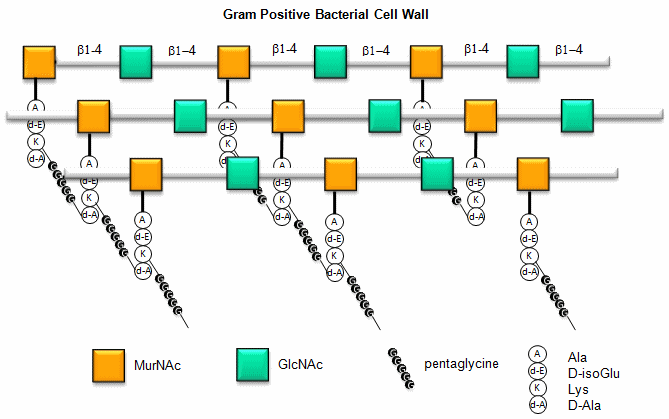
Here is a figure which give a better view of the overall cell wall of Gram-positive bacteria.
Carbohydrates in biomolecule recognition
Carbohydrates are covalently attached to many different biomolecules, including lipids, to form glycolipids, and proteins, to form glycoproteins. Glycoproteins and glycolipids are often found in biological membranes, to which they are anchored by through nonpolar interactions. A special kind of glycoprotein, a proteoglycan, actually has more carbohydrate mass than protein. What is the function of these carbohydrates? Two are apparent. First, glycosylation of proteins helps protect the protein from degradation by enzyme catalysts within the body. However, there main functions arises from the fact that covalently attached carbohydrates that "decorate" the surface of glycoproteins or glycolipids provide new binding site interactions that allow interactions with other biomolecules. Hence glycosylation allows for cell:cell, cell:protein, or protein:protein interactions. Unfortunately, bacteria and viruses often recognize glycosylated molecules on cell membranes, allowing for their import into the cell.
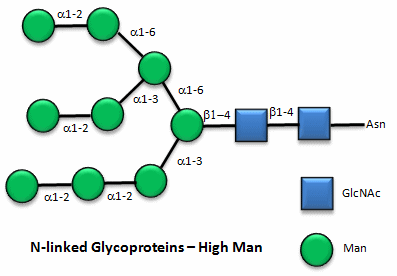
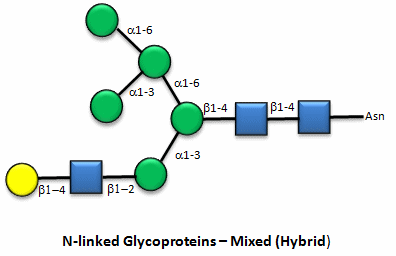
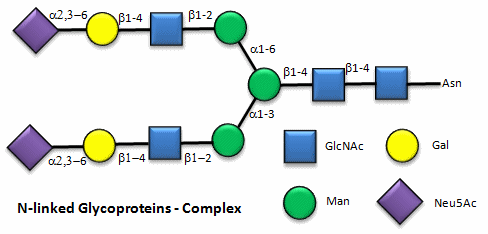
Here are some "cartoon" examples of carbohydrates covalently linked to the amino acid asparagine (Asn) on a glycoprotein.
Here are some examples of biomolecular interactions promoted by IMFs involving carbohydrates.
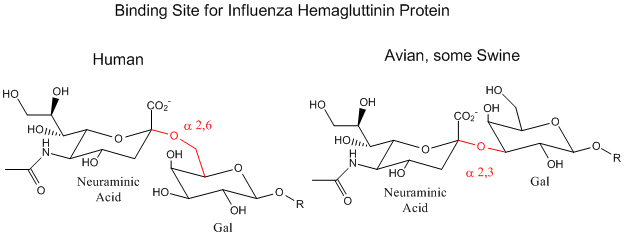
Influenza Virus binding to Cell Surface Glycoproteins with Neu5Ac - A protein on the surface of influenza virus, hemagluttinin, bind to sialic acid (Sia), which is covalently attached to many cell membrane glycoproteins on host cells. The sialic acid is usually connected through an alpha (2,3) or alpha (2,6) link to galactose on N-linked glycoproteins. The subtypes found in avian (and equine) influenza isolates bind preferentially to Sia (alpha 2,3) Gal which predominates in avian GI tract where viruses replicate. Human virus of H1, H2, and H3 subtype (cause of the 1918, 1957, and 1968 pandemics) recognize Sia (alpha 2,6) Gal, the major form in human respiratory tract. The swine influenza HA bind to Sia (alpha 2,6) Gal and some Sia (alpha 2,3)both of which found in swine.
- Jmol model of viral hemagluttinin bound to antiviral drugs and sialic (neuraminic acid) from Proteopedia
Leukocyte: Cell Wall binding - During inflammation, circulating leukocytes (a type of white blood cell) tether and roll on the walls of blood vessels where they become active. E-, L- and P-selectin proteins are the primary proteins responsible for the tethering and rolling of these leukocytes. P-selectin binds, in part, to a tetrasaccharide, sialyl-Lewisx (SLEX) on the cell surface.. The interaction between P-selectin and the cell mediates the initial binding/rolling of the leukocyte on the vessel wall.