1.8: Intermolecular forces
- Page ID
- 225767
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Dipoles
To understand the nature of noncovalent interactions, we first must return to covalent bonds and delve into the subject of dipoles. Many of the covalent bonds that we have seen – between two carbons, for example, or between a carbon and a hydrogen –involve the approximately equal sharing of electrons between the two atoms in the bond. In these examples, the two atoms have approximately the same electronegativity. Recall from your general chemistry course that electronegativity refers to “ the power of an atom in a molecule to attract electrons to itself” (this is the definition offered by Linus Pauling, the eminent 20th-century American chemist who was primarily responsible for developing many of the bonding concepts that we have been learning).
However, quite often in organic chemistry we deal with covalent bonds between two atoms with different electronegativities, and in these cases the sharing of electrons is not equal: the more electronegative nucleus pulls the two electrons closer. In the carbon-oxygen bond of an alcohol, for example, the two electrons in the sigma bond are held more closely to the oxygen than they are to the carbon, because oxygen is significantly more electronegative than carbon. The same is true for the oxygen-hydrogen bond, as hydrogen is slightly less electronegative than carbon, and much less electronegative than oxygen.
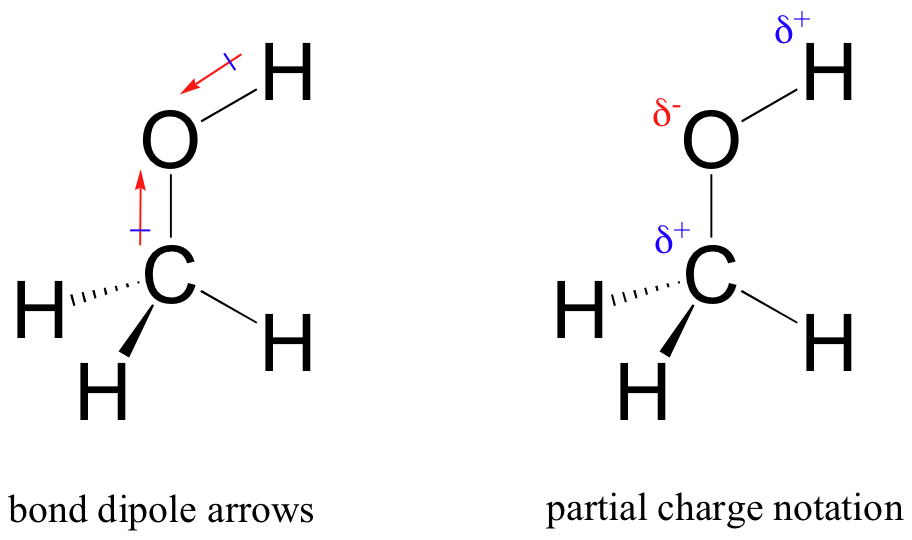
The result of this unequal sharing is what we call a bond dipole, which exists in a polar covalent bond. A bond dipole has both negative and positive ends, or poles, where electron density is lower (the positive pole) and higher (the negative pole). The difference in electron density can be expressed using the Greek letter delta to denote ‘partial positive’ and ‘partial negative’ charge on the atoms. ‘Dipole arrows’, with a positive sign on the tail, are also used to indicated the negative (higher electron density) direction of the dipole.
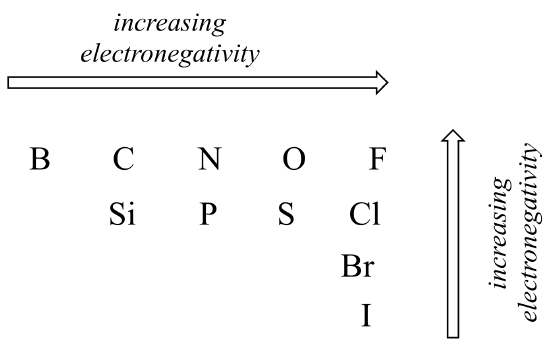
The degree of polarity in a covalent bond depends on the difference in electronegativity between the two atoms. Electronegativity is a periodic trend: it increases going from left to right across a row of the periodic table of the elements, and also increases as we move up a column. Therefore, oxygen is more electronegative than nitrogen, which is in turn more electronegative than carbon. Oxygen is also more electronegative than sulfur. Fluorine, in the top right corner of the periodic table, is the most electronegative of the elements. Hydrogen is slightly less electronegative than carbon. For a full table of electronegativity values, see section 1.3.
Periodic trends in electronegativity
Exercise
Using what you about atomic orbitals, rationalize the periodic trends in electronegativity. Why does it increase from left to right, and decrease from top to bottom? This is a good question to talk through with classmates and an instructor or tutor.
[reveal-answer q=”811978″]Show Solution[/reveal-answer]
[hidden-answer a=”811978″]
The horizontal trend is based on atomic number (the number of protons in the nucleus). For example, fluorine is more electronegative than carbon, because the fluorine nucleus contains three more protons, the positive charges on which pull negatively-charged electrons closer to the nucleus.
The vertical trend is based on atom size, specifically the size of the ‘electron cloud’ surrounding the nucleus. For example, fluorine is more electronegative than chlorine (even though chlorine contains more protons) because the outermost valence electrons on fluorine, which are in the n = 2 “shell”, are closer to the nucleus than the valence electrons in chlorine, which occupy the n = 3 “shell”. The fluorine electron cloud, therefore, is subject to greater electrostatic attractive forces from protons (electrostatic forces decrease rapidly as the distance between the positive and negative charges increases.)
[/hidden-answer]
Most molecules contain both polar and nonpolar covalent bonds. Depending on the location of polar bonds and bonding geometry, molecules may posses a net polarity, called a molecular dipole moment. Water, as you probably recall, has a dipole moment that results from the combined dipoles of its two oxygen-hydrogen bonds. Fluoromethane also has a dipole moment.

Tetrafluoromethane, however, has four polar bonds that pull equally in to the four corners of a tetahedron, meaning that although there are four bond dipoles there is no overall molecular dipole moment. Carbon dioxide also lacks a molecular dipole moment.
Exercise
Which of the molecules below have molecular dipole moments?
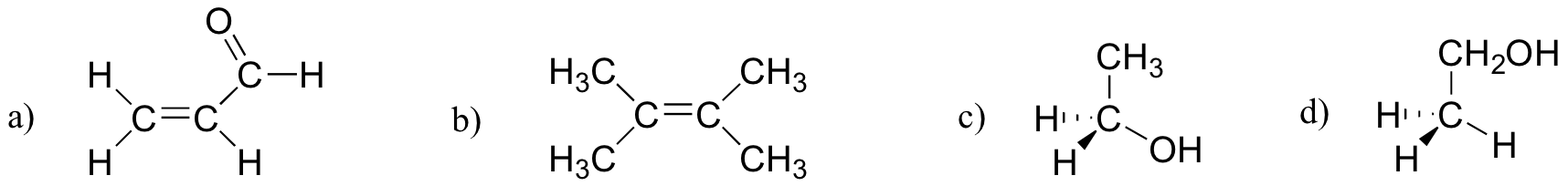
[reveal-answer q=”636950″]Show Solution[/reveal-answer]
[hidden-answer a=”636950″]
Only molecule (b) does not have a molecular dipole, due to its symmetry (bond dipoles are equal and in opposite directions).[/hidden-answer]
Ion-ion, dipole-dipole and ion-dipole interactions
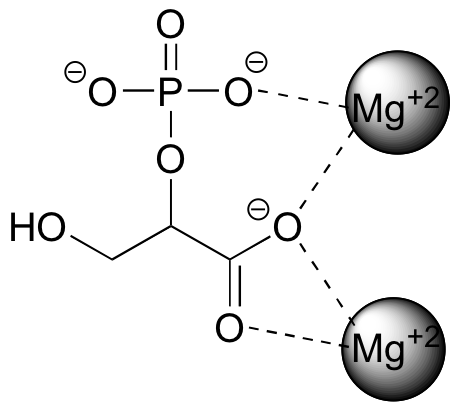
The strongest type of non-covalent interaction is between two ionic groups of opposite charge (an ion-ion or charge-charge interaction). You probably saw lots of examples of ionic bonds in inorganic compounds in your general chemistry course: for example, table salt is composed of sodium cations and chloride anions, held in a crystal lattice by ion-ion interactions. One of the most common examples in biological organic chemistry is the interaction between a magnesium cation (Mg+2) and an anionic carboxylate or phosphate group. The figure below shows 2-phosphoglycerate, an intermediate in the glycolysis pathway, interacting with two Mg+2 ions in the active site of a glycolytic enzyme called enolase.
Polar molecules – those with an overall dipole moment, such as acetone – can align themselves in such a way as to allow their respective positive and negative poles to interact with each other. This is called a dipole-dipole interaction.

When a charged species (an ion) interacts favorably with a polar molecule or functional group, the result is called an ion-dipole interaction. A common example of ion-dipole interaction in biological organic chemistry is that between a metal cation, most often Mg+2 or Zn+2, and the partially negative oxygen of a carbonyl.

Because the metal cation is very electronegative, this interaction has the effect of pulling electron density in the carbonyl double bond even further toward the oxygen side, increasing the partial positive charge on carbon. As we shall later, this has important implications in terms of the reactivity of carbonyl groups in biochemical reactions.
Van der Waals forces
Nonpolar molecules such as hydrocarbons also are subject to relatively weak but still significant attractive noncovalent forces. Van der Waals forces (also called London dispersion forces or nonpolar interactions) result from the constantly shifting electron density in any molecule. Even a nonpolar molecule will, at any given moment, have a weak, short-lived dipole. This transient dipole will induce a neighboring nonpolar molecule to develop a corresponding transient dipole of its own, with the end result that a transient dipole-dipole interaction is formed. These van der Waals forces are relatively weak, but are constantly forming and dissipating among closely-packed nonpolar molecules, and when added up the cumulative effect can become significant.
Hydrogen bonds
Hydrogen bonds result from the interaction between a hydrogen bonded to an electronegative heteroatom – specifically a nitrogen, oxygen, or fluorine – and lone-pair electrons on a nitrogen, oxygen, or fluorine a neighboring molecule or functional group. Because a hydrogen atom is just a single proton and a single electron, when it loses electron density in a polar bond it essentially becomes an approximation of a ‘naked’ proton, capable of forming a strong interaction with a lone pair on a neighboring electronegative atom.

Hydrogen bonds are usually depicted with dotted lines in chemical structures. A group that provides a proton to a hydrogen bond is said to be acting as a hydrogen bond donor. A group that provides an oxygen or nitrogen lone pair is said to be acting as a hydrogen bond acceptor. Many common organic functional groups can participate in the formation of hydrogen bonds, either as donors, acceptors, or both. Water and alcohols, for example, can be both hydrogen bond donors and acceptors. A carbonyl, as it lacks a hydrogen bound to an oxygen or nitrogen, can only act as a hydrogen bond acceptor.
Exercise
Classify the structures below as:
A) capable of being both a hydrogen bond donor and acceptor
B) capable of being a hydrogen bond acceptor, but not a donor
C) not capable of participating in hydrogen bonding.

[reveal-answer q=”44735″]Show Solution[/reveal-answer]
[hidden-answer a=”44735″]
To be a hydrogen bond donor, the molecule needs to have a hydrogen bound to N, O, or F. To be an acceptor, it merely needs an N, O, or F.

[/hidden-answer]
Exercise
Draw figures that show the hydrogen bonds described below.
a) A hydrogen bond between methanol (donor) and water (acceptor).
b) A hydrogen bond between methanol (acceptor) and water (donor).
c) Two possible hydrogen bonds between methyl acetate and methylamine.
[reveal-answer q=”787121″]Show Solution[/reveal-answer]
[hidden-answer a=”787121″]
Note in part (c) that methyl acetate can only be a hydrogen bond acceptor, not a donor.
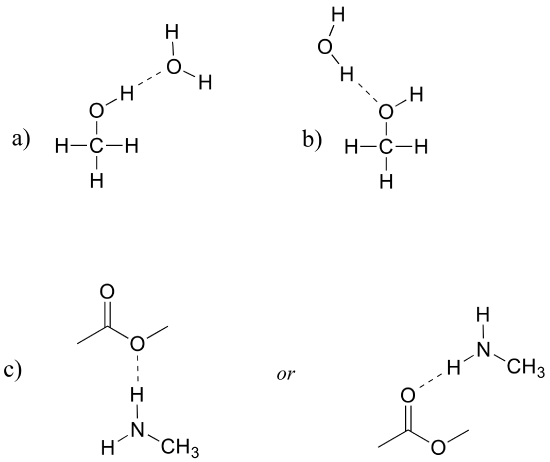
[/hidden-answer]
In general, hydrogen bonds are stronger than dipole-dipole interactions, but also much weaker than covalent bonds. The strength of hydrogen bonds has enormous implications in biology. Copying of DNA in the cell, for example, is based on very specific hydrogen bonding arrangements between DNA bases on complimentary strands: adenine pairs with thymine, while guanine pairs with cytidine:

Hydrogen bonds, as well as the other types of noncovalent interactions, are very important in terms of the binding of a ligand to a protein. In section 1.3, we saw a ‘space-filling’ picture of an enzyme with its substrate bound in its active site. Here, in a two-dimensional approximation, is an image of the same substrate-enzyme pair showing how amino acid side chain (green) and parent chain (blue) groups surround and interact with functional groups on the substrate (red).


- Organic Chemistry With a Biological Emphasis. Authored by: Tim Soderbergu00a0(University of Minnesota, Morris). Located at: https://chem.libretexts.org/Textbook_Maps/Organic_Chemistry/Book%3A_Organic_Chemistry_with_a_Biological_Emphasis_(Soderberg)/Chapter_02%3A_Introduction_to_organic_structure_and_bonding_II/2.4%3A_Non-covalent_interactions. Project: Chemistry LibreTexts. License: CC BY-NC-SA: Attribution-NonCommercial-ShareAlike


