4.1: PubChem Web Interfaces for Text
- Page ID
- 170159
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)PubChem Homepage
The PubChem homepage (https://pubchem.ncbi.nlm.nih.gov) provides a search interface that allow users to perform any term/keyword/identifier search against all three major databases of PubChem1,2,3: Compound, Substance, BioAssay. If a search returns multiple hits, they are presented on an Entrez DocSum page and will be explained in more detail later in this chapter. If the search returns a single record, the user will be directed to the web page that presents information on that record. This page is called the Compound Summary, Substance Record, or BioAssay Record page, depending on the record type (i.e., compound, substance, or assay). In addition, the PubChem homepage provides launch points to various PubChem services, tools, help documents, and more. In general, the PubChem homepage is a central location for all PubChem services.
Entrez Search and Retrieval System
NCBI’s Entrez4,5,6,7 is a database retrieval system that integrates PubChem’s three major databases as well as other NCBI’s major databases, including PubMed, Nucleotide and Protein Sequences, Protein Structures, Genome, Taxonomy, BioSystems, Gene Expression Omnibus (GEO) and many others. Entrez provides users with an integrated view of biomedical data and their relationships. This section focuses on search and retrieval of PubChem data using the Entrez system. A more detailed description on the Entrez system is given in the following documents:
- The Entrez Search and Retrieval System
(http://www.ncbi.nlm.nih.gov/books/NBK184582/) - Entrez Help
(https://www.ncbi.nlm.nih.gov/books/NBK3836/)
Entry points to Entrez
One can search the PubChem databases through Entrez, by initiating a search from the NCBI home page (http://www.ncbi.nlm.nih.gov). By default, if a specific database is not selected in the search menu, Entrez searches all Entrez databases available, and lists the number of records in each database that are returned for this “global query”. The following link directs you to the global query result page for the term “AIDS” against all databases integrated in the Entrez system.
https://www.ncbi.nlm.nih.gov/gquery/?term=AIDS
Simply by selecting one of the three PubChem databases from the global query results page (under the Chemical section), one can see the query results specific to that database.
Alternatively, one can start from the PubChem home page (http://pubchem.ncbi.nlm.nih.gov), where a search of one of the three PubChem databases may be initiated through the search box at the top. It is also possible to initiate an Entrez search against a PubChem database from the following pages:
- https://www.ncbi.nlm.nih.gov/pccompound/ (to search the Compound database)
- https://www.ncbi.nlm.nih.gov/pcsubstance/ (to search the Substance database)
- https://www.ncbi.nlm.nih.gov/pcassay/ (to search the BioAssay database)
Entrez DocSums
If an Entrez search for a query against any of the three PubChem databases returns a single record, the user will be directed to the Compound Summary, Substance Record, or BioAssay Record page for that record (depending on whether the record is a compound, substance, or assay). If it returns multiple records, Entrez will display a document summary report (also called “DocSum” page). The following link directs you to the DocSum page for a search for the term “lipitor” against the PubChem Compound database:
https://www.ncbi.nlm.nih.gov/pccompound?term=lipitor
In this example, the DocSum page displays a list of the compound records returned from the search. For each record, some data-specific information is provided with a link to the summary page for that record. The DocSum page contains controls to change the display type, to sort the results by various means, or to export the page to a file or printer. Additional controls that operate on a query result list are available on the right column of the DocSum page. The DocSum page for the other two PubChem databases look similar to this example for the Compound database.
Entrez Indices
Entrez indices, tied to individual records in an Entrez database, include information on particular aspects (often referred to as fields) of the records. These indices may have text, numeric or date values, and some indices may have multiple values for each record. The available fields and their indexed terms in any Entrez database can be found from the drop-down menus on the Advanced Search Builder page (which can be accessed by clicking the “Advanced” link next to the “Go” button on the PubChem Home page).
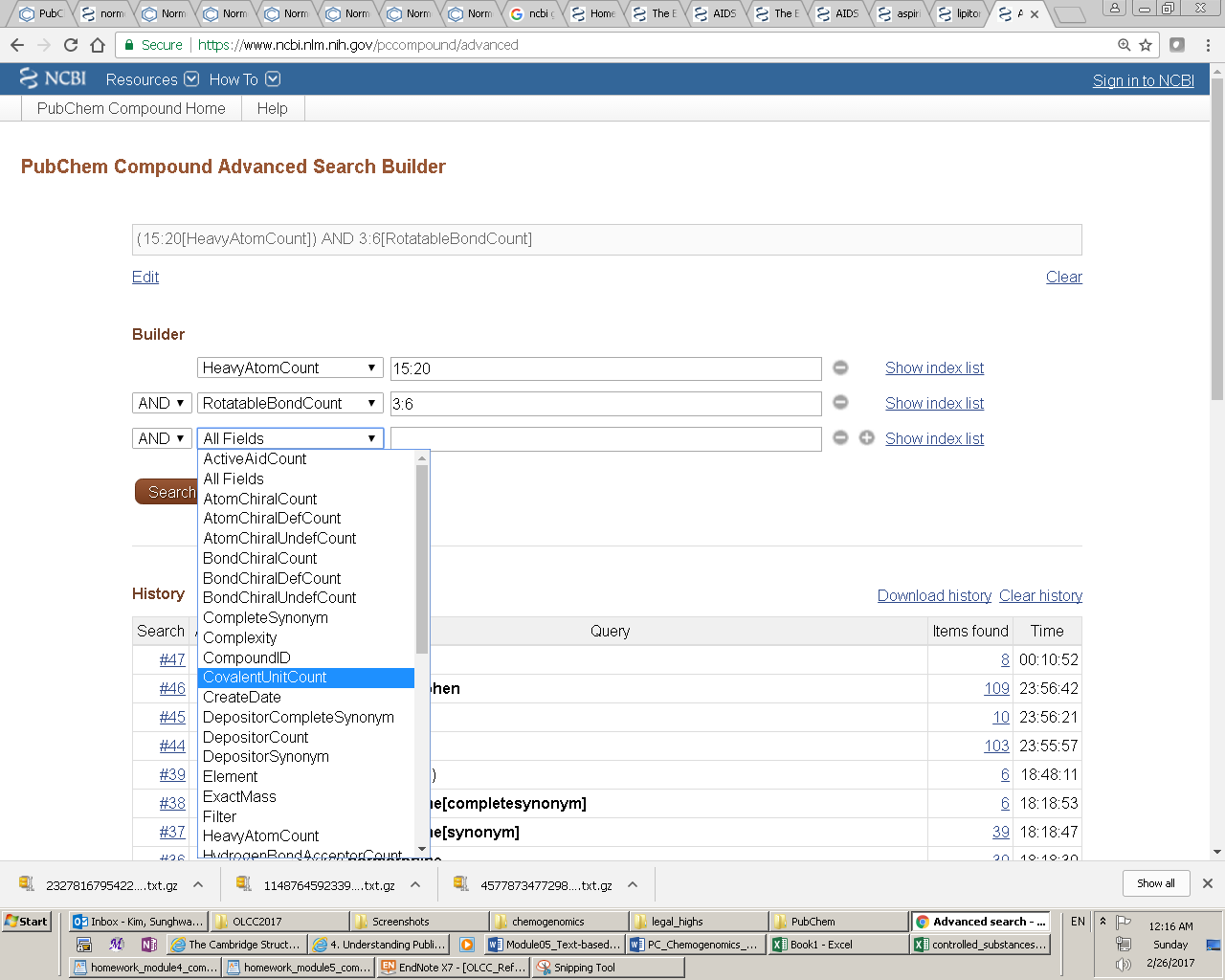
When the user enters a query in the Entrez search interface, the Entrez indices are matched directly to that query. By default, in an Entrez search with a simple query, all indexed fields are matched against the query, usually resulting in the largest number of returned records including many unwanted results. One can narrow the search to a particular indexed field, by adding the index name in brackets after the term itself (e.g., “lipitor[synonym]”). For numeric indices, a search for a range of values can be done by using minimum and maximum values separated by a colon and followed by the bracketed index name (e.g., “100:105[MolecularWeight]”). Multiple indices may be searched simultaneously using Entrez’s Boolean operators (e.g., “AND”, “OR” and “NOT”).
A complete list of the Entrez indices available for the three PubChem databases can be retrieved in the XML format, using the eInfo functionality in E-Utilities (which will be covered in Module 7):
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pccompound (for Compound)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcsubstance (for Substance)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcassay (for BioAssay).
Additional information on the PubChem Entrez indices is available in the “Indices and Filters in Entrez” section of the help documentation:
https://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Index
Entrez Links
Entrez links are cross links or associations between records in different Entrez databases, or within the same database. These links may be applied to an entire search result list (via the “find related data” section at the right column of a DocSum page) or to an individual record (via links at the bottom of each record presented on the DocSum page). The Entrez links provide a way to discover relevant information in other Entrez databases based on a user’s specific interests. Equivalently, one may think of this as a way to transform an identifier list from one database to another based on a particular criterion. Note that there are limits to how many records may be used as input in a link operation. To process a large amount of input records and/or to expect a large amount of output records associated with the input records, one should use the FLink tool (https://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi).
A complete list of the Entrez links available for the three PubChem databases can be retrieved in the XML format through these links
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pccompound (for Compound)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcsubstance (for Substance)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcassay (for BioAssay).
Entrez Filters
Entrez filters are essentially Boolean bits (true or false) for all records in a database that indicate whether or not a given record has a particular property. The Entrez filters may be used to subset other Entrez searches according to this property, by adding the filter to the query string.
Entrez filters are closely related to links in that the majority of Entrez filters in the PubChem databases are generated automatically based on whether PubChem records have Entrez links to a given database. However, some special filters, such as the "lipinski rule of 5" filter, or the “all” filter, are not link-based.
The Entrez filters available for each Entrez database may be found on the Advanced Search Builder page by selecting “Filter” from the “All Fields” dropdown and clicking “Show index list”.
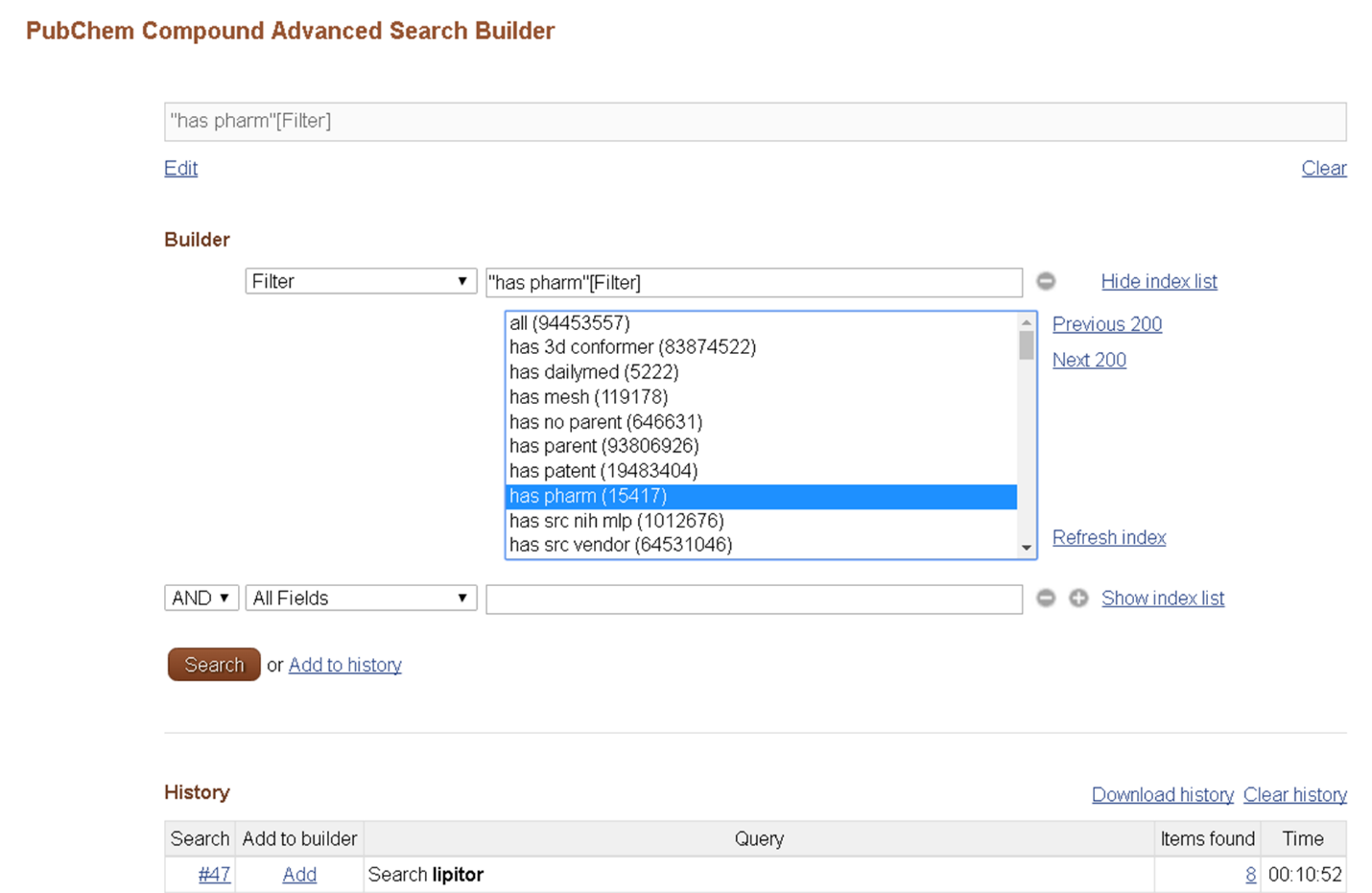
More detailed description of the Entrez filters available for the three PubChem databases are given in the “Indices and Filters in Entrez” section of the help documentation:
https://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Index
Entrez History
Entrez has a history mechanism (Entrez history) that automatically keeps track of a user’s searches, temporarily caches them (for eight hours), and allows one to combine search result sets with Boolean logic (i.e., “AND”, “OR”, and “NOT”). The Entrez history allows one to limit a search to a subset of records returned from a previous search. Use of Entrez history can help users avoid sending and receiving (potentially) very large lists of identifiers. In addition, through the Entrez history, one can use the search results as an input to various PubChem tools for further manipulation and analysis.
References
(3) Kim, S. Expert Opinion on Drug Discovery 2016, 11, 843.
(4) Schuler, G. D.; Epstein, J. A.; Ohkawa, H.; Kans, J. A. Methods Enzymol. 1996, 266, 141.
(5) McEntyre, J. Trends in genetics : TIG 1998, 14, 39.
(6) The Entrez Search and Retrieval System (https://www.ncbi.nlm.nih.gov/books/NBK184582/) (Accessed on.
(7) Entrez Help (https://www.ncbi.nlm.nih.gov/books/NBK3836/) (Accessed on.

