2.6: Chemical Resolvers, Molecular Editors and Visualization
- Page ID
- 154858
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Introduction
Chemical resolvers can take one form of molecular representation and convert it to another. That is, they can resolve what the compound is from its represenation. They can be web based services or software applications. A molecular editor is in essence a type of resolver, that has a graphical editor interface where human's can draw molecules. But under the hood, it is using cheminformatics representations like connection tables. Database services like PubChem and ChemSpider also have integrated editors and resolvers and so the distinction across these is a bit fuzzy.
Chemical Resolvers
We will define chemical resolvers as programs that can resolve a chemical structure from a representation, and then use that to transform it to another representation or provide information on the chemical.
Web-Based Resolvers
These are services that typically offer both a GUI (for humans) and an API (for machines). This list is not comprehensive, and will grow as time allows.
CIR
The Chemical Identifier Resolver (CIR) is a service of the Computer-Aided Drug Design (CADD) group of the Chemical Biology Laboratory (CBL) of the National Cancer Institute (NCI) in Maryland US. The direct link to CIR is here:
https://cactus.nci.nih.gov/chemical/structure
Figure \(\PageIndex{1}\): The GUI interface for the NCI/CADD CIR (https://cactus.nci.nih.gov/chemical/structure).
The CIR service also offers a variety of API interfaces and we already explored one of them with the InChILayersExplorer of activity 2.4.1 (section 2.4.3.3.1), where an Excel spreadsheet used the CIR service to convert a name to an InChI.
There are also a variety of other resources available through the CADD Group Chemiformatics Tools and User Services (CACTUS) that students are encouraged to explore.
OPSIN
The Open Parser for Systematic IUPAC Nomenclature (OPSIN) is run by the Centre for Molecular Informatics and the University of Cambridge in England, the URL is:
Figure \(\PageIndex{2}\): OPSIN resolver, (https://opsin.ch.cam.ac.uk/)
One of the nice things of the OPSIN resolver is that if you have an incorrect IUPAC name, it stops where it can't parse the name and tell's you where the problem is. For example, patents will often use IUPAC names and believe it or not, they are often misspelled and wrong! So lets look at this name here,
1-[4-(2-methoxyethy)phenoxy]-3-(propan-2-amino)propan-2-ol.
Do you see what is wrong with it? Well pasting it into OPSIN gives you an error and an idea where to look!

Figure \(\PageIndex{3}\): OPSIN resolver showing an error in resolving the IUPAC name.
The beauty is the error message occurs when the resolver could no longer parse the word and it got stumped at the y), which should have been yl), as in 1-[4-(2-methoxyethyl)phenoxy]-3-(propan-2-ylamino)propan-2-ol.
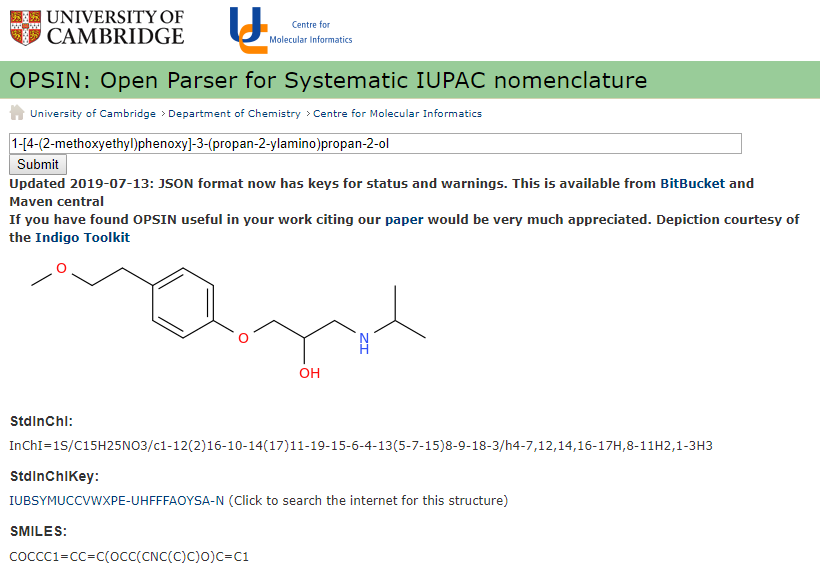
Figure \(\PageIndex{4}\): Discovering the type in figure 3 allowed us to correct the IUPAC name and we now have identifiers for this compound.
The above figures show how the OPSIN GUI can be used. In activity X we will look at a Google Spreadsheet that uses the OPSIN resolver to convert a list of IUPAC names to InChIKeys and then use those keys to link directly to PubChem on those chemicals, and thus instantly get access to information on a list of compounds. Lets manually do that now. That is, we append the InChI key to following URL stem
https://pubchem.ncbi.nlm.nih.gov/compound/INCHIKEY
and so the following link gets us to information on this compound
https://pubchem.ncbi.nlm.nih.gov/compound/IUBSYMUCCVWXPE-UHFFFAOYSA-N
bringing us to the following information on PubChem
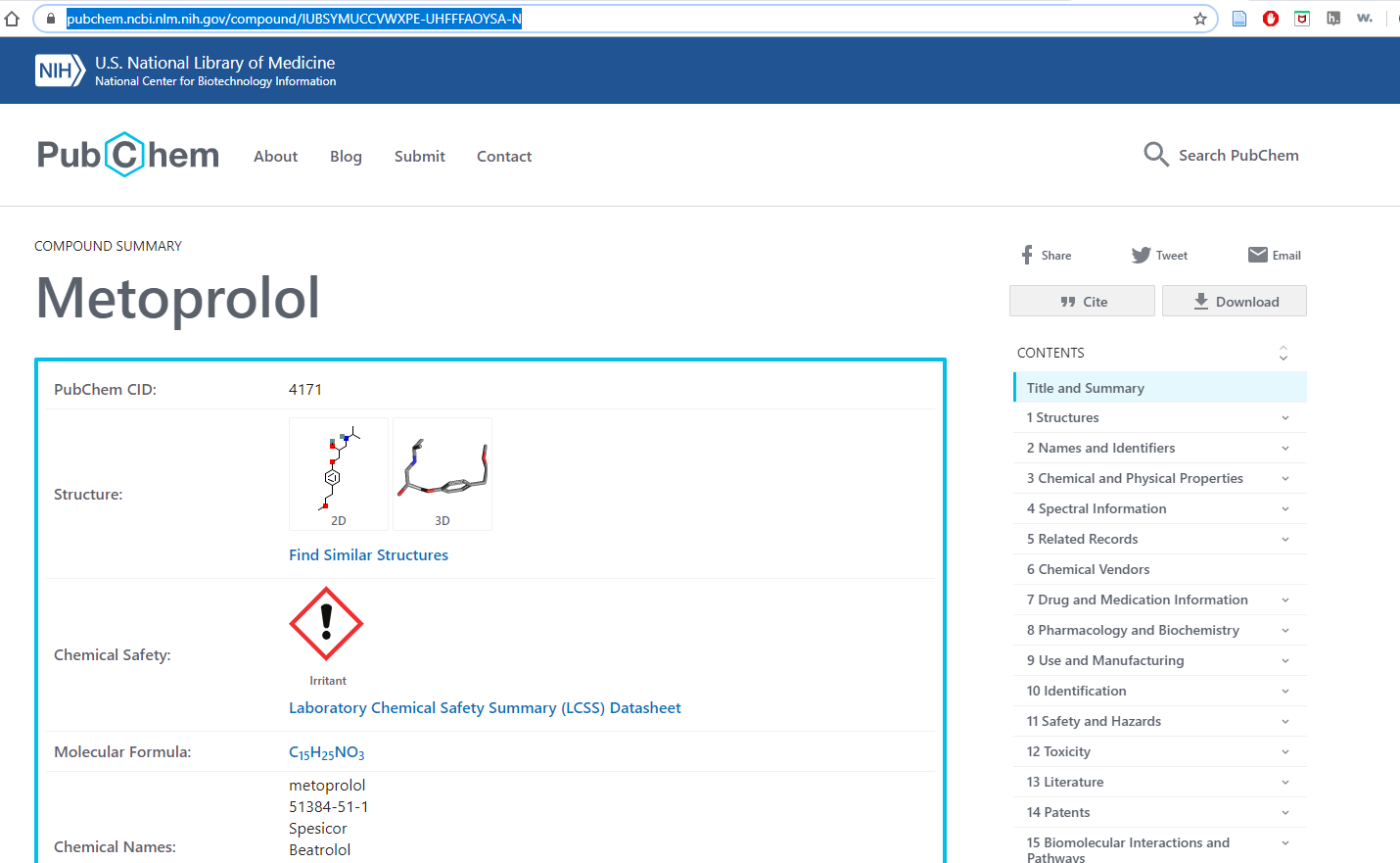
Figure \(\PageIndex{5}\): After using OPSIN to correct the incorrect IUPAC name we can deduce the compound is Metoprolol and obtain information on it.
IUPAC Name2PubChem
Go to the InChI OER (https://www.inchi-trust.org/oer/) and filter Content Type/Spreadsheet with File Type/Google Sheet, and click on the hit for the "IUPAC Name2PubChem". From here you will find a Google sheet that automates the above process for a list of chemicals. Video \(\PageIndex{1}\) shows how you can go to the OPSIN chemical resolver and figure out how to create a Google sheet that performs these functions.
Video \(\PageIndex{1}\): 6:47 min YouTube video describing how to connect an column of IUPAC names to information in PubChem (https://youtu.be/oDxMUJ0dNWw)
If you download the module from the InChI OER you will see the code described in the video which you can cut and paste into a spreadsheet. This module uses two webservices functions, the IMPORTDATA function and the HYPERLINK function, along with the concatenate function for a string of text, &. It is very important that you develop the skill of looking at code you have never seen and then finding out what it does through web searches.
Resolver Programs
OK, Resolver Programs may not be the best title for this section. These are software packages that convert molecular file formats, and are often called tool kits. These have many functionalities that can be used offline, in contrast to web services, which need internet access.
Open Babel
Open Babel is an open source Chemistry Toolbox and does much more than just convert structural formats and can be downloaded from the following URL.
In figure \(\PageIndex{6}\) we see using Open Babel to convert a SMILES string to a mol file with 3D coordinates. The Open Babel Documentation can give you a feel for some of the things you can do with Open Babel, http://openbabel.org/docs/current/OpenBabel.pdf .
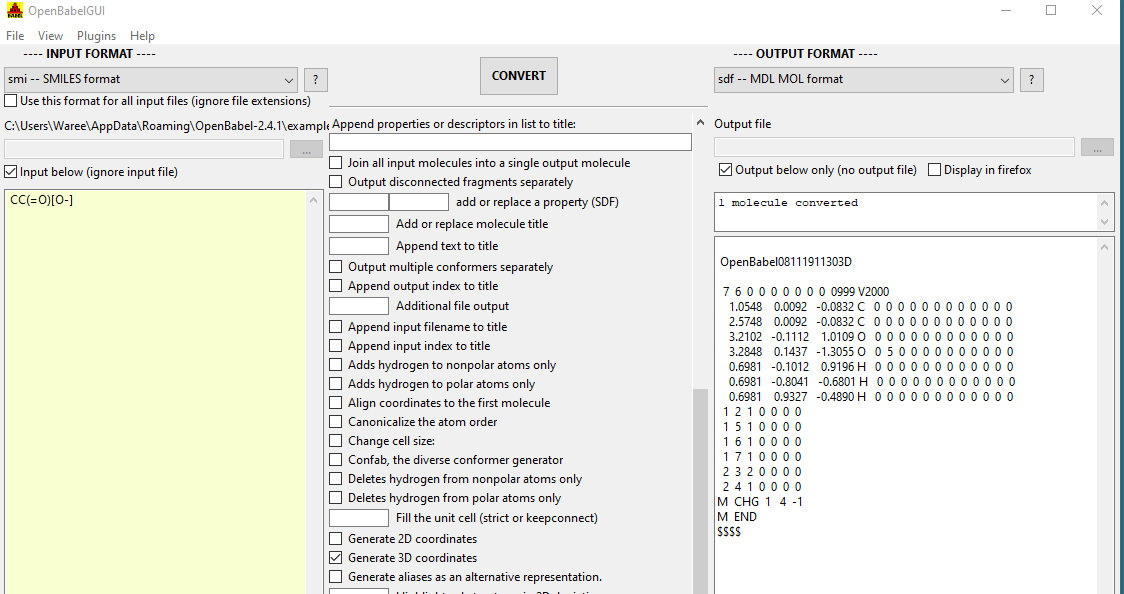
Figure \(\PageIndex{6}\): Using Open Babel to convert between file types, here converting a SMILES string to a 3D mol file with coordinates.
Molecular Editors
Web Services
- MolView - site maintained by Herman Bergwerf
- PubChem Sketcher
- ChemSpider Structure Search
- Chemagic
- Wikipedia Chemical Structure Explorer
- http://www.cheminfo.org/Wikipedia/
- see JChem Inf. article (https://jcheminf.biomedcentral.com/articles/10.1186/s13321-015-0061-y)
Software Packages
Dr. Tamas Gunda has posted a decent resource: Chemical Drawing Programs - The Comparison of Accelrys (Biovia) Draw, ChemBioDraw (ChemDraw), DrawIt, ChemDoodle and Chemistry 4-D Draw (http://www.gunda.hu/dprogs/index.html)
Open Source
- JChemPaint - open source (LGLP license):
- BIOVIA Draw – free for academic use
- ChemSketch Freeware - free for academic and personal use, requires registration and download:
- BIO-RAD - Chemical Structure drawing, spectral analysis & more
- JSME - open source (BSD license):
Fee-Based
- ChemDraw – requires subscription and download
- Basic drawing package: http://www.cambridgesoft.com/Ensemble_for_Chemistry/ChemDraw/ChemDrawPrime/Default.aspx
- ChemDoodle – requires purchase and download
- ChemSketch – requires purchase and download
- http://www.acdlabs.com/products/draw_nom/draw/chemsketch/
- Free Trial at above link
- Chemistry 4-D Draw
Molecular Visualization
- Jmol and Jsmol
- Avogadro

