4.3: DNA Replication
- Page ID
- 347426
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Source: BiochemFFA_7_2.pdf. The entire textbook is available for free from the authors at http://biochem.science.oregonstate.edu/content/biochemistry-free-and-easy
Copying instructions
The only way to make new cells is by the division of pre-existing cells. Single-celled organisms undergo division to produce more cells like themselves, while multicellular organisms arise through division of a single cell, generally the fertilized egg. Each time a cell divides, all of its DNA must be copied faithfully so that a copy of this information can be passed on to the daughter cell. This process is called DNA replication. It is the means by which genetic information can be transmitted down generations of cells, and it ensures that every new cell has a complete copy of the genome. In the next section, we will examine the process by which the DNA of a cell is completely and accurately copied.
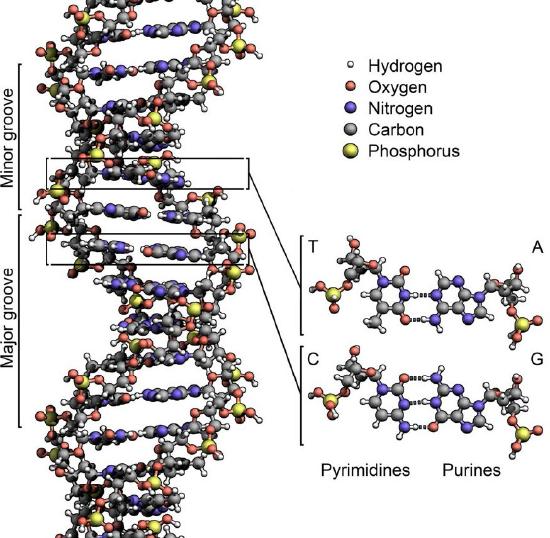
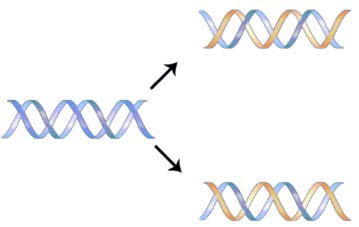
The structure of DNA elucidated by Watson and Crick in 1953 immediately suggested a mechanism by which double-stranded DNA could be copied to give two identical copies of the DNA. They proposed that the two strands of the DNA molecule, which are held together by hydrogen bonds between the base-paired nucleotides, would separate and each serve as a template on which a complementary strand could be assembled (Figure 7.8). The base-pairing rules would ensure that this process would result in the production of two identical DNA molecules. The beautiful simplicity of this scheme was shown to be correct in subsequent experiments by Meselson and Stahl, that demonstrated that DNA replication was semi-conservative, i.e., that after replication, each of the two resulting DNA molecules was made up of one old strand and one new strand that had been assembled across from it (Figure 7.9).
Building materials
What are the ingredients necessary for building a new DNA molecule? As noted above, the original, or parental DNA molecule serves as the template. New DNA molecules are assembled across from each template by joining together free DNA nucleotides as directed by the base pairing rules, with As across from Ts and Gs across from Cs.
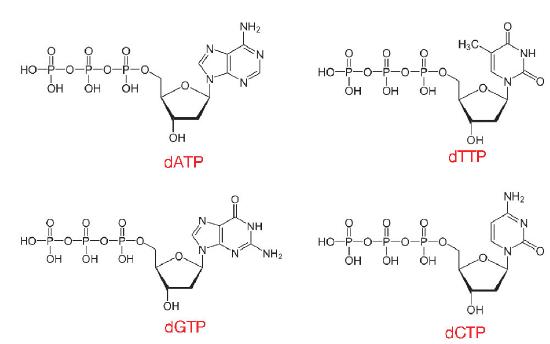
The nucleotides used in DNA synthesis are deoxyribonucleoside triphosphates or dNTPs. As can be inferred from their name, such nucleotides have a deoxyribose sugar and three phosphates, in addition to one of the four DNA bases, A, T, C or G (Figure 7.10).
When dNTPs are added into a growing DNA strand, two of those phosphates will be cleaved off, as described later, leaving the nucleotides in a DNA molecule with only one phosphate per nucleotide. This reaction is catalyzed by enzymes known as DNA polymerases, which create phosphodiester linkages between one nucleotide and the next.
Challenges
Before examining the actual process of DNA replication, it is useful to think about what it takes to accomplish this task successfully. Consider the challenges facing a cell in this process:
- The sheer number of nucleotides to be copied is enormous: e.g., in human cells, on the order of several billions.
- A double-helical parental DNA molecule must be unwound to expose single strands of DNA that can serve as templates for the synthesis of new DNA strands.
- Unwinding must be accomplished without introducing topological distortion into the molecule.
- The unwound single strands of DNA must be kept from coming back together long enough for the new strands to be synthesized.
- DNA polymerases cannot begin synthesis of a new DNA strand de novo and require a free 3' OH to which they can add deoxynucleotides.
- DNA polymerases can only extend a strand in the 5' to 3' direction. The 5' to 3' growth of both new strands means that one of the strands is made in pieces.
- The use of RNA primers requires that the RNA nucleotides must be removed and replaced with DNA nucleotides and the resulting DNA fragments must be joined.
- The copying of all the parental DNA must be accurate, so that mutations are not introduced into the newly made DNA.
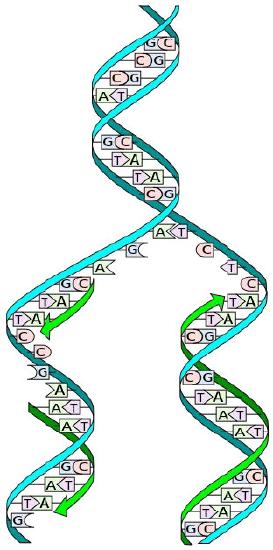

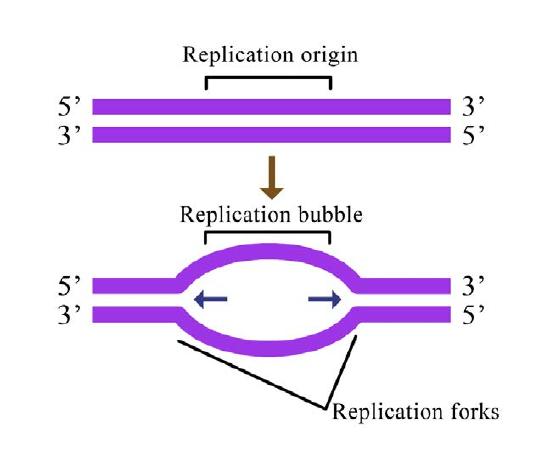
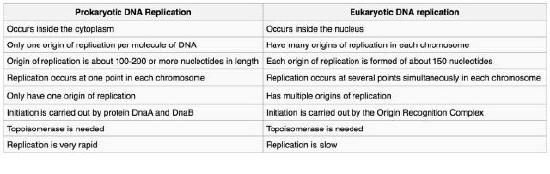
Figure 7.14 - Prokaryotic vs. eukaryotic DNA replication - Wikipedia
Addressing challenges
With this in mind, we can begin to examine how cells deal with each of these challenges. Our understanding of the process of DNA replication is derived from studies using bacteria, yeast, and other systems. These investigations have revealed that DNA replication is carried out by the action of a large number of proteins that act together as a complex protein machine. Numerous proteins involved in replication have been identified and characterized, including multiple different DNA polymerases in both prokaryotes and eukaryotes. Although the specific proteins involved are different in bacteria and eukaryotes, it is useful to understand the basic considerations that are relevant in all cells. A generalized account of the steps in DNA replication is presented below, focused on the challenges mentioned above.
- The sheer number of nucleotides to be copied is enormous: e.g., in human cells, on the order of several billions.
Cells, whether bacterial or eukaryotic, have to replicate all of their DNA before they can divide. In cells like our own, the vast amount of DNA is broken up into many chromosomes, each of which is composed of a linear strand of DNA (Figure 7.12). In cells like those of E. coli, there is a single circular chromosome.
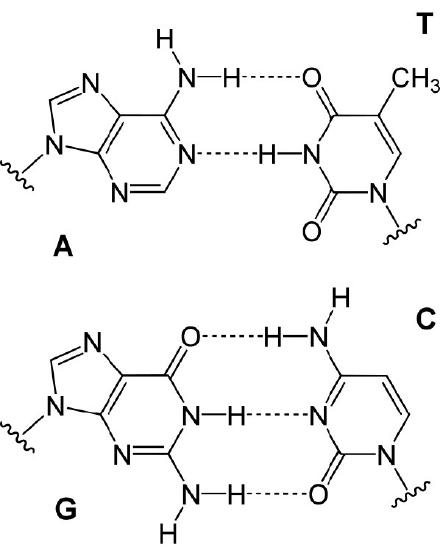
In either situation, DNA replication is initiated at sites called origins of replication. These are regions of the DNA molecule that are recognized by special proteins called initiator proteins that bind the DNA. In E.coli, origins have small regions of A-T-rich sequences that are “melted” to separate the strands, when the initiator proteins bind to the origin or replication. As you may remember, A-T base-pairs, which have two hydrogen bonds between them are more readily disrupted than G-C base-pairs which have three apiece (Figure 7.15).
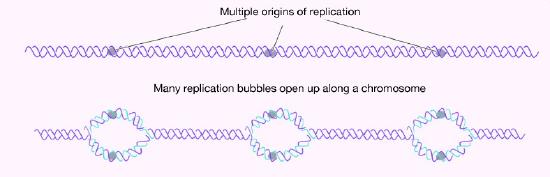
How many origins of replication are there on a chromosome? In the case of E. coli, there is a single origin of replication on its circular chromosome. In eukaryotic cells there may be many thousands of origins of replication, with each chromosome having hundreds (Figure 7.16). DNA replication is, thus, initiated at multiple points along each chromosome in eukaryotes. Electron micrographs of replicating DNA from eukaryotic cells show many replication bubbles on a single chromosome. This makes sense in light of the large amount of DNA that there is to be copied in cells like our own, where beginning at one end of each chromosome and replicating all the way through to the other end from a single origin would simply take too long. This is despite the fact that the DNA polymerases in human cells are capable of building new DNA strands at the very respectable rate of about 50 nucleotides per second!
- A double-helical parental molecule must be unwound to expose single strands of DNA that can serve as templates for the synthesis of new DNA strands.
Unwinding
Once a small region of the DNA is opened up at each origin of replication, the DNA helix must be unwound to allow replication to proceed. The unwinding of the DNA helix requires the action of an enzyme called helicase.
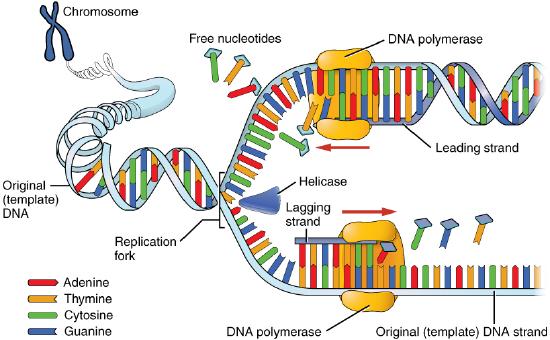
Helicase uses the energy released when ATP is hydrolyzed, to break the hydrogen bonds between the bases in DNA and separate the two strands (Figure 7.17). Note that a replication bubble is made up of two replication forks that "move" or open up, in opposite directions. At each replication fork, the parental DNA strands must be unwound to expose new sections of single-stranded template.
- This unwinding must be accomplished without introducing topological distortion into the molecule.
What is the effect of unwinding one region of the double helix? Local unwinding of the double helix causes over-winding (increased positive supercoiling) ahead of the unwound region.
The DNA ahead of the replication fork has to rotate, or it will get twisted on itself and halt replication. This is a major problem, not only for circular bacterial chromosomes, but also for linear eukaryotic chromosomes, which, in principle, could rotate to relieve the stress caused by the increased supercoiling.
Topoisomerases
The reason this is problematic is that it is not possible to rotate the entire length of a chromosome, with its millions of base-pairs, as the DNA at the replication fork is unwound. How, then, is this problem solved? Enzymes called topoisomerases can relieve the topological stress caused by local “unwinding” of the extra winds of the double helix. They do this by cutting one or both strands of the DNA and allowing the strands to swivel around each other to release the tension before rejoining the ends. In E. coli, the topoisomerase that performs this function is called gyrase.
- The separated single strands of DNA must be kept from coming back together so the new strands to be synthesized.
Single-strand DNA binding protein
Once the two strands of the parental DNA molecule are separated, they must be prevented from going back together to form double-stranded DNA. To ensure that unwound regions of the parental DNA remain single-stranded and available for copying, the separated strands of the parental DNA are bound by many molecules of a protein called single-strand DNA binding protein (SSB - Figure 7.18).
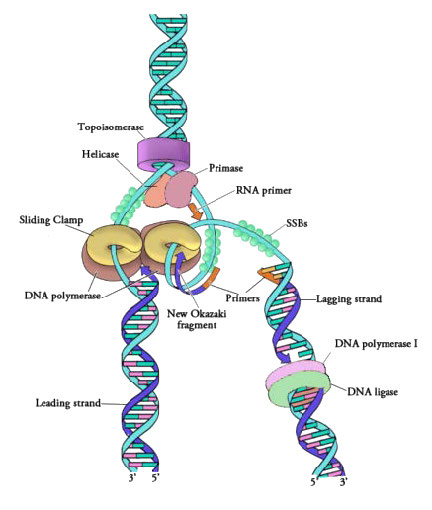
Figure 7.18 - Proteins at a prokaryotic DNA replication fork - Image by Martha Baker
- DNA polymerases cannot begin synthesis of a new DNA strand de novo and require a free 3' OH to which they can add DNA nucleotides.
Although single-stranded parental DNA is now available for copying, DNA polymerases cannot begin synthesis of a complementary strand de novo. This simply means that DNA polymerases can only add new nucleotides on to the 3' end of a pre-existing chain, and cannot start a chain of nucleotides on their own. Because of this limitation, some enzyme other than a DNA polymerase must first make a small region of nucleic acid, complementary to the parental strand, that can provide a free 3' OH to which DNA polymerase can add a deoxyribonucleotide. This task is accomplished by an enzyme called a primase, which assembles a short stretch of RNA base-paired to the parental DNA template. This provides a short base-paired region, called the RNA primer, with a free 3'OH group to which DNA polymerase can add the first new DNA nucleotide (Figure 7.12).
Sliding clamp
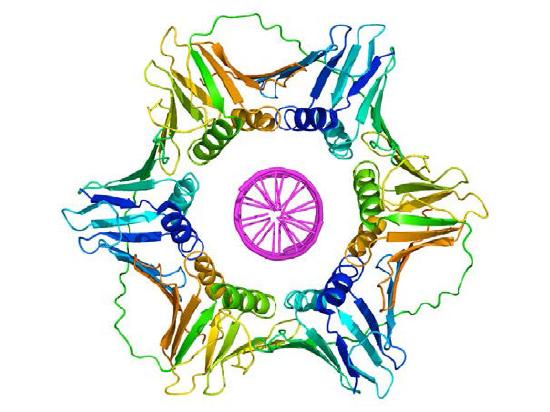
Once a primer provides a free 3'OH for extension, other proteins get into the act. These proteins are involved in loading the DNA polymerase onto the primed template and keeping it associated with the DNA. The first of these is the clamp loader. As its name suggests, the clamp loader helps to load a protein complex called the sliding clamp onto the DNA at the replication fork (Figure 7.19 and 7.20). The sliding clamp, a multi-subunit ring-shaped protein, is then joined by the DNA Polymerase. The function of the sliding clamp is to keep the polymerase associated with the replication fork - in fact, it has been described as a seat-belt for the DNA polymerase. The sliding clamp ensures that the DNA polymerase is able to synthesize long stretches of new DNA before it dissociates from the template. The property of staying associated with the template for a long time before dissociating is known as the processivity of the enzyme. In the presence of the sliding clamp, DNA polymerases are much more processive, making replication faster and more efficient.
Extending the primer
The DNA polymerase is now poised to start synthesis of the new DNA strand (in E. coli, the primary replicative polymerase is called DNA polymerase III). As you already know, the synthesis of new DNA is accomplished by the addition of new nucleotides complementary to those on the parental strand. DNA polymerase catalyzes the reaction by which an incoming deoxyribonucleotide, complementary to the template, is added onto the 3' end of the previous nucleotide, starting with the 3'OH on the end of the RNA primer. The importance of the 3’OH group lies in the nature of the reaction that builds a chain of nucleotides.
The reaction catalyzed by the DNA polymerase occurs through the nucleophilic attack by the 3’OH group at the end of a nucleic acid strand on the α phosphate of the incoming dNTP (Figure 7.21). The immediate hydrolysis of the pyrophosphate that is cleaved off the incoming dNTP drives the reaction forward. The sequential addition of new nucleotides at the 3’ end of the growing chain of DNA accounts for the fact that the strand grows in a 5’ to 3’ direction.
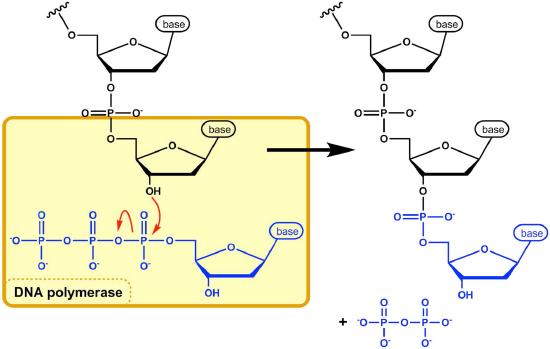
The 5' phosphate on each incoming nucleotide is joined by the DNA polymerase to the 3' OH on the end of the growing nucleic acid chain, to make a phosphodiester bond. Each added nucleotide provides a new 3’OH, allowing the chain to be extended for as long as the DNA polymerase continues to synthesize the new strand. As we already noted, the new DNA strands are synthesized by the addition of DNA nucleotides to the end of an RNA primer. The new DNA molecule thus has a short piece of RNA at the beginning.
- DNA polymerases can only extend a strand in the 5' to 3' direction. The 5' to 3' growth of both new strands means that one of the strands is made in pieces.
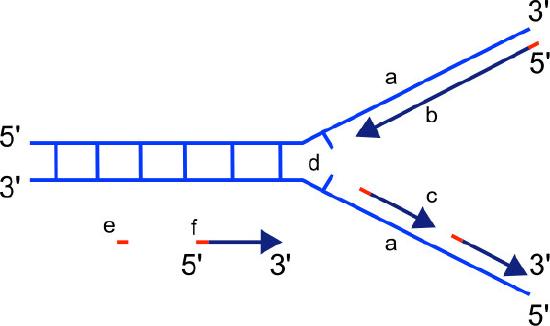
Leading strand
We know that DNA polymerases can only build a new DNA strand in the 5' to 3' direction. We also know that the two parental strands of DNA are antiparallel. This means that at each replication fork, one new strand, called the leading strand can be synthesized continuously in the 5' to 3' direction because it is being made in the same direction that the replication fork is opening up.
Lagging strand
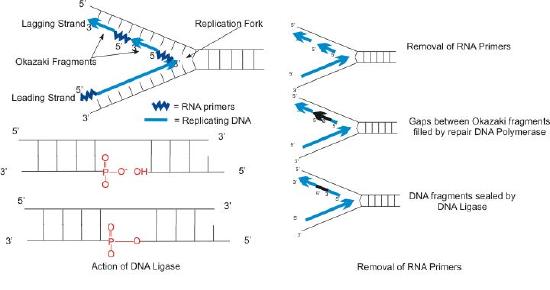
The synthesis of the other new strand, called the lagging strand, also proceeds in the 5’ to 3’ direction. But because the template strands are running in opposite directions, the lagging strand is being extended in the direction opposite to the opening of the replication fork (Figure 7.22). As the replication fork opens up, the region behind the original start point for the lagging strand will need to be copied. This means another RNA primer must be laid down and extended. This process repeats itself as the replication fork opens up, with multiple RNA primers laid down and extended, producing many short pieces that are later joined. These short nucleic acid pieces, each composed of a small stretch of RNA primer and about 1000-2000 DNA nucleotides, are called Okazaki fragments, for Reiji Okazaki, the scientist who first demonstrated their existence.
- The use of RNA primers requires that the RNA nucleotides must be removed and replaced with DNA nucleotides.
Primer removal
We have seen that each newly synthesized piece of DNA starts out with an RNA primer, effectively making a new nucleic acid strand that is part RNA and part DNA. The newly made DNA strand cannot be allowed to have pieces of RNA attached. So, the RNA nucleotides must be removed and the gaps filled in with DNA nucleotides (Figure 7.23). This is done by DNA polymerase I in E. coli. This enzyme begins adding DNA nucleotides at the end of each Okazaki fragment. However, the end of one Okazaki fragment is adjacent to the RNA primer at the beginning of the next Okazaki fragment. DNA polymerase I has an exonuclease activity acting in the 5’ to 3’ direction that removes the RNA nucleotides ahead of it, while the polymerase activity replaces the RNA nucleotides with dNTPs. Once all the RNA nucleotides have been removed, the lagging strand is made up of stretches of DNA. The DNA pieces are then joined together by the enzyme DNA ligase.
The steps outlined above essentially complete the process of DNA replication. But one issue still remains.
- Ensuring accuracy in the copying of so much information
Accuracy
How accurate is the copying of information by DNA polymerase? As you are aware, changes in DNA sequence (mutations) can change the amino acid sequence of the encoded proteins and that this is often, though not always, deleterious to the functioning of the organism. When billions of bases in DNA are copied during replication, how do cells ensure that the newly synthesized DNA is a faithful copy of the original information?
DNA polymerases, as we have noted earlier work fast (averaging 50 bases a second in human cells and up to 200 times faster in E. coli). Yet, both human and bacterial cells seem to replicate their DNA quite accurately. This is because replicative DNA polymerases have a proofreading function that enables the polymerase to detect when the wrong base has been inserted across from a template strand, back up and remove the mistakenly inserted base, before continuing with synthesis (Figure 7.24).
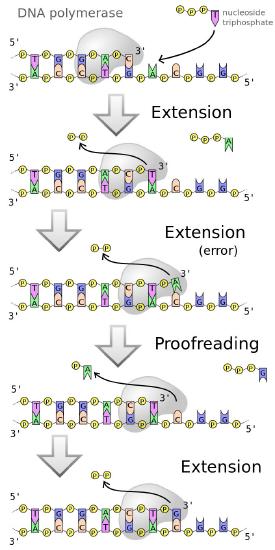
Figure 7.24 - Error corrected by DNA polymerases
Multiple activities
This is possible because most DNA polymerases are dual-function enzymes. They can extend a DNA chain by virtue of their 5' to 3' polymerase activity. Some polymerases like DNA polymerase I can also remove RNA primers in the 5’ to 3’ direction, though that is not a common activity of polymerases. Many polymerases, however have an ability to backtrack and remove the last inserted base because they possess a 3' to 5' exonuclease activity.
The exonuclease activity of a DNA polymerase allows it to excise a wrongly inserted base, after which the polymerase activity inserts the correct base and proceeds with extending the strand.
In other words, the DNA polymerase is monitoring its own accuracy (also termed its fidelity) as it makes new DNA, correcting mistakes immediately before moving on to add the next base. This mechanism, which operates during DNA replication, corrects many errors as they occur, reducing by about a 100-fold the mistakes made when DNA is copied.
DNA polymerases
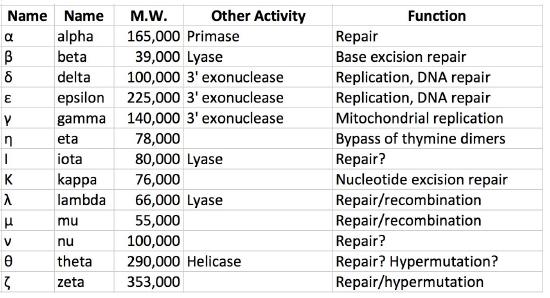
As noted earlier, both prokaryotic and eukaryotic cells have multiple DNA polymerases. In E.coli, for example, DNA polymerase III is the major replicative polymerase (a.k.a. replicase) while DNA polymerase I is responsible for DNA repair as well as removal of RNA primers and their replacement with DNA nucleotides during replication. DNA polymerase II plays a role in restarting replication after DNA damage stalls replication, while DNA polymerases IV and V are both required in trans-lesion, or bypass, synthesis, which allows replication past sites of DNA damage.
Eukaryotic polymerases
In eukaryotes, there are over fifteen different DNA polymerases. The primary replicative polymerases in the nucleus are ∂ and ε. DNA polymerase α is also important for replication because it has primase and repair activities. Replication is initiated in eukaryotic cells by DNA polymerase α, which binds to the initiation complex at the origin and lays down an RNA primer, followed by about 25 nucleotides of DNA. It is then replaced by another polymerase, in a step called the pol switch. DNA polymerase ∂ or ε then continues synthesizing DNA, depending on the strand. The role of polymerase ε appears to be synthesis of the leading strand due to its high processivity and accuracy, whereas polymerase ∂ extends Okazaki fragments on the lagging strands. Proteins analogous to the clamp loader and sliding clamp are also present. The protein RFC plays the role of clamp loader, while another protein, PCNA acts like the sliding clamp. Several other DNA polymerases like β, γ and μ function in repairing gaps. Yet others are involved in trans-lesion synthesis following DNA damage and are associated with hypermutation.
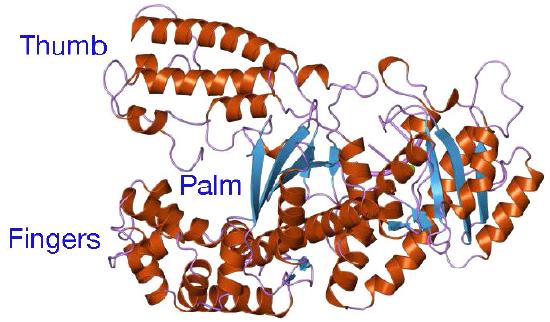
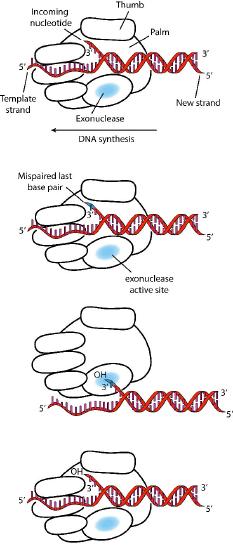
Despite their diversity, DNA polymerases share some common structural features. X-ray crystallographic studies have shown that these enzymes have a structure that has been compared to a human right hand (Figures 7.25 & 7.26). The «palm» of the hand forms a cleft in which the DNA lies. The cleft is also the where the polymerase catalytic activity resides. This is where the incoming nucleotide is added on to the growing chain. «The fingers» position the DNA in the active site, while the «thumb» holds the DNA as it exits the polymerase. A separate domain contains the exonuclease (proofreading) activity of the enzyme.
The enzyme alternates between its polymerizing activity and its proofreading activity. When a mismatched base pair is in the polymerase catalytic site, the 3’end of the growing strand is moved from the polymerase site to the exonuclease active site (Figure 7.26). The mispair at the end is removed by the exonuclease, followed by repositioning of the 3’ end in the polymerase active site to continue synthesis.
Termination of replication
In circular bacterial chromosomes, there are specific sequences known as terminator or Ter sites. These are multiple short sequences that serve as termination sites, allowing the replication forks traveling clockwise and anticlockwise across the circular chromosome to meet at one of the sites.
The binding of a protein, Tus, at a Ter site prevents further movement of the replication fork and ends replication. The parental and newly made circular DNA are, at this point topologically interlinked and must be separated with the help of topoisomerase.
The end-replication problem
There is no fixed site for termination in linear eukaryotic chromosomes. As the replication forks reach the ends of the chromosome, the leading strand can be synthesized all the way to the end of the template strand. On the lagging strand, the need for an RNA primer to start synthesis creates a challenge. When the RNA primer at the extreme end of the lagging strand is removed, there is a small stretch of the template strand that cannot be copied. As a result, in each round of replication a short sequence at the ends of the chromosome will be lost. Over time, with many cycles of replication, chromosomes would become noticeably shorter. This shortening of chromosomes has been observed in vitro, in cultured mammalian somatic cells. It is also seen in intact organisms, with increasing age.
Telomeres
What effect does the loss of sequence from the ends of the chromosomes have on cells? We know that the ends of chromosomes are characterized by structures called telomeres (Figure 7.28). Telomeres are made up of many copies of short repeated sequences (in humans, the repeat is TTAGGG) and special proteins that specifically bind to these sequences. This structure of telomeres is useful in distinguishing the ends of chromosomes from double-strand breaks in DNA, thus preventing the DNA repair mechanisms in cells from joining chromosomes end to end.
The other advantage of the repeated sequences, which do not encode proteins, is that losing some of the repeats does not lead to loss of important coding information. Thus, the repeats act as a sort of buffer zone, where the loss of sequence does not doom the cell. However, the shortening of chromosomes cannot continue indefinitely. After a certain number of replication cycles, cells are known to stop dividing and enter a state known as replicative senescence. This suggests that the shortening of the telomeres serves as a sort of clock, with the extent of shrinkage of the chromosomes serving as a measure of aging. Eventually cells that enter senescence will die.
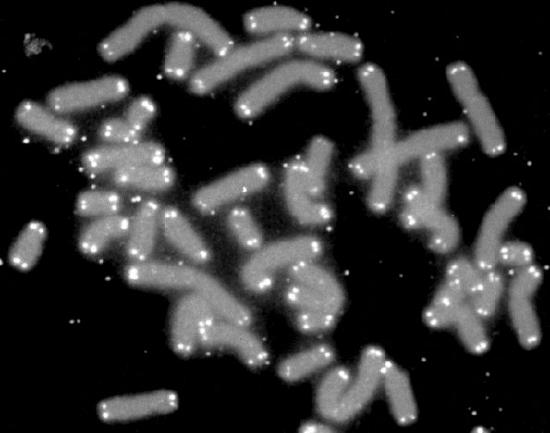
Figure 7.28 - Chromosomes with telomeres marked in white
Problems with sequence loss
Even if our cells are able to function with shorter chromosomes during our lifetimes, this leaves us with another problem. If our chromosomes grow shorter with age, then presumably our children, who inherit our chromosomes will be born with shorter chromosomes than we started with. They, in turn, would have their chromosomes shrink as they grew older, and their children would have even shorter chromosomes. Over the course of multiple generations, this would lead to the point where further chromosome shrinkage would result in cells that would enter senescence very early in life and die soon after. This obviously does not happen. Generation after generation of children are born with full-length chromosomes, so there is a mechanism that must ensure that at least in the reproductive cells, chromosomes do not get shorter.
To understand this mechanism, it is necessary to first examine the end of a newly made DNA molecule (Figure 7.29). While the leading strand, which grows in the same direction as the movement of the replication fork, can copy its template all the way to the end, the lagging strand encounters a problem. RNA primers are, as we noted, needed to start each of the Okazaki fragments of the lagging strand. The primers must be removed later, and the RNA nucleotides replaced with DNA nucleotides. When the RNA primer across from the end of the parental strand is removed, the RNA nucleotides cannot be replaced by DNA nucleotides because the DNA polymerase has no primer to start from. A short region of the template cannot, therefore be copied.

Figure 7.29 - Replication of a linear chromosome results in loss of sequences at the very ends with each round of replication
Telomerase
How can this problem be solved? It can be seen from Figure 7.29 that the end of the original template strand has a short 3’ overhang resulting from the removal of the RNA primer across from it. In order to fill in this region, another primer would be needed, situated past the end of the template strand. But in order to build such a primer, it would be necessary for the template overhang to be longer. If it were possible to make the template strand longer, then another primer could be placed across from its end and the end of the strand could be copied. Such an extension of the template strand is exactly what happens in our reproductive cells. The parental template strand is extended by the enzyme telomerase, which adds telomere repeats and lengthens the template. We will see shortly how it accomplishes that feat.
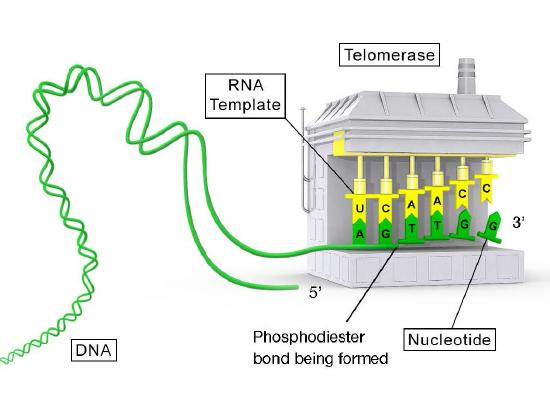
RNA template
Telomerase is an unusual enzyme, in that it is made up of two components, an RNA and a reverse transcriptase. A reverse transcriptase is an RNA-dependent DNA polymerase, an enzyme that copies an RNA template to make DNA. The RNA component of the human telomerase, called hTERC, has a sequence that is complementary to the telomere repeat, TAGGG. As seen in Figure 7.31, this RNA can base-pair with the last telomere repeat on the parental DNA strand, while a portion of the RNA remains unpaired.
Template for extension
The function of the unpaired region of the RNA is to serve as a template that can be used to extend the overhanging 3’ end of the original DNA molecule. The protein component of telomerase has reverse transcriptase activity and can copy the RNA sequence into DNA. In human telomerase, the protein component is known as hTERT (telomerase reverse transcriptase). As seen in Figure 7.31 and 7.32, the reverse transcriptase extends the original 3’ overhang using the RNA component as its template. The telomerase can then dissociate, and repeat the process multiple times to add many repeats of the telomere sequence.
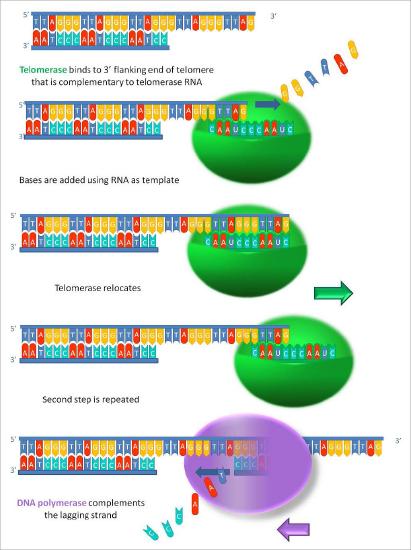
Once the overhang has been extended by the addition of at least several telomere repeats, there is now room for the synthesis of an RNA primer complementary to the newly extended overhang (pointing back towards the rest of the chromosome). This primer can then be extended to complete synthesis of the lagging strand all the way to the end of the original parental DNA strand. Thus, the addition of telomere repeats on the parental DNA strands keeps the newly made DNA strands from becoming shorter with each cycle of replication. The fact that this happens in germ cells (reproductive cells) explains why each generation does not have shorter chromosomes than the parental generation.
The proofreading function of DNA polymerases monitors the accuracy of DNA replication while the enzyme telomerase keeps chromosomes that will be passed on to offspring from shortening. Between them, these two activities ensure that the genetic information is copied accurately, and that succeeding generations receive a full complement of the genetic information
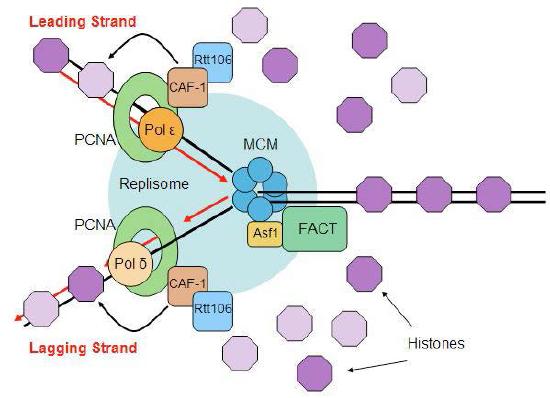
Disassembly and reassembly of nucleosomes
The events of replication have an additional twist in eukaryotes. Recall that DNA is found in eukaryotic cells as chromatin, a complex of the DNA with proteins. At its least condensed, chromatin looks like a string of beads, consisting of the DNA wrapped around histone cores to make nucleosomes. The nucleosome structure must be disrupted to make DNA available for replication and restored after replication is completed (Figure 7.33).
Ahead of the replication fork, chromatin structure is disassembled by ATP-dependent chromatin remodeling complexes, allowing access to the DNA template. Once the new strands of DNA have been synthesized, both the original nucleosomes and new nucleosomes must be reassembled behind the replication fork. Since replication gives rise to two DNA molecules where there was one, twice the amount of histones is needed to package the DNA. Preparation for DNA replication, therefore, involves the synthesis of large amounts of histones to supply the need. Interestingly, it appears that newly synthesized DNA is packaged into nucleosomes using the original histones that were displaced to allow the replication fork to pass, as well as newly synthesized histones.
We also know that post-translational modifications like acetylation, methylation or phosphorylation of the histones can regulate the degree to which a given region of the genome is accessible for use. One question that remains the subject of intense research is how these modifications are accurately passed on to the new nucleosomes.