The transition metals and zinc are among the least abundant metal ions in the sea water from which contemporary organisms are thought to have evolved (Table 1.1).1-5 For many of the metals, the concentration in human blood plasma greatly exceeds that in sea water. Such data indicate the importance of mechanisms for accumulation, storage, and transport of transition metals and zinc in living organisms.
Table 1.1: Concentrations of transition metals and zinc in sea water and human plasma.a*Data from References 1 - 5 and 12
| Element |
Sea Water (M) x 108 |
Human Plasma (M) x 108 |
| Fe |
0.005 - 2 |
2230 |
| Zn |
8.0 |
1720 |
| Cu |
1.0 |
1650 |
| Mo |
10.0 |
1000 |
| Co |
0.7 |
0.0025 |
| Cr |
0.4 |
5.5 |
| V |
4.0 |
17.7 |
| Mn |
0.7 |
10.9 |
| Ni |
0.5 |
4.4 |
The metals are generally found either bound directly to proteins or in cofactors such as porphyrins or cobalamins, or in clusters that are in turn bound by the protein; the ligands are usually O, N, S, or C. Proteins with which transition metals and zinc are most commonly associated catalyze the intramolecular or intermolecular rearrangement of electrons. Although the redox properties of the metals are important in many of the reactions, in others the metal appears to contribute to the structure of the active state, e.g., zinc in the Cu-Zn dismutases and some of the iron in the photosynthetic reaction center. Sometimes equivalent reactions are catalyzed by proteins with different metal centers; the metal binding sites and proteins have evolved separately for each type of metal center.
Iron is the most common transition metal in biology.6,7 Its use has created a dependence that has survived the appearance of dioxygen in the atmosphere ca. 2.5 billion years ago, and the concomitant conversion of ferrous ion to ferric ion and insoluble rust (Figure 1.1 See color plate section, page C-1.). All plants, animals, and bacteria use iron, except for a lactobacillus that appears to maintain high concentrations of manganese instead of iron. The processes and reactions in which iron participates are crucial to the survival of terrestrial organisms, and include ribonucleotide reduction (DNA synthesis), energy production (respiration), energy conversion (photosynthesis), nitrogen reduction, oxygen transport (respiration, muscle contraction), and oxygenation (e.g., steroid synthesis, solubilization and detoxification of aromatic compounds). Among the transition metals used in living organisms, iron is the most abundant in the environment. Whether this fact alone explains the biological predominance of iron or whether specific features of iron chemistry contribute is not clear.
Many of the other transition metals participate in reactions equivalent to those involving iron, and can sometimes substitute for iron, albeit less effectively, in natural Fe-proteins. Additional biological reactions are unique to nonferrous transition metals.
Zinc is relatively abundant in biological materials.8,9 The major location of zinc in the body is metallothionein, which also binds copper, chromium, mercury, and other metals. Among the other well-characterized zinc proteins are the Cu-Zn superoxide dismutases (other forms have Fe or Mn), carbonic anhydrase (an abundant protein in red blood cells responsible for maintaining the pH of the blood), alcohol dehydrogenase, and a variety of hydrolases involved in the metabolism of sugars, proteins, and nucleic acids. Zinc is a common element in nucleic-acid polymerases and transcription factors, where its role is considered to be structural rather than catalytic. Interestingly, zinc enhances the stereoselectivity of the polymerization of nucleotides under reaction conditions designed to simulate the environment for prebiotic reactions. Recently a group of nucleic-acid binding proteins, with a repeated sequence containing the amino acids cysteine and histidine, were shown to bind as many as eleven zinc atoms necessary for protein function (transcribing DNA to RNA).10 Zinc plays a structural role, forming the peptide into multiple domains or "zinc fingers" by means of coordination to cysteine and histidine (Figure 1.2A See color plate section, page C-l.). A survey of the sequences of many nucleic-acid binding proteins shows that many of them have the common motif required to form zinc fingers. Other zinc-finger proteins called steroid receptors bind both steroids such as progesterone and the progesterone gene DNA (Chapter 8). Much of the zinc in animals and plants has no known function, but it may be maintaining the structures of proteins that activate and deactivate genes.11
Copper and iron proteins participate in many of the same biological reactions:
- reversible binding of dioxygen, e.g., hemocyanin (Cu), hemerythrin (Fe), and hemoglobin (Fe);
- activation of dioxygen, e.g., dopamine hydroxylase (Cu) (important in the synthesis of the hormone epinephrine), tyrosinases (Cu), and catechol dioxygenases (Fe);
- electron transfer, e.g., plastocyanins (Cu), ferredoxins, and c-type cytochromes (Fe);
- dismutation of superoxide by Cu or Fe as the redox-active metal (superoxide dismutases).
The two metal ions also function in concert in proteins such as cytochrome oxidase, which catalyzes the transfer of four electrons to dioxygen to form water during respiration. Whether any types of biological reactions are unique to copper proteins is not clear. However, use of stored iron is reduced by copper deficiency, which suggests that iron metabolism may depend on copper proteins, such as the serum protein ceruloplasmin, which can function as a ferroxidase, and the cellular protein ascorbic acid oxidase, which also is a ferrireductase.
Cobalt is found in vitamin B12 , its only apparent biological site.12 The vitamin is a cyano complex, but a methyl or methylene group replaces CN in native enzymes. Vitamin-B12 deficiency causes the severe disease of pernicious anemia in humans, which indicates the critical role of cobalt. The most common type of reaction in which cobalamin enzymes participate results in the reciprocal exchange of hydrogen atoms if they are on adjacent carbon atoms, yet not with hydrogen in solvent water:
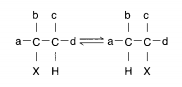
(An important exception is the ribonucleotide reductase from some bacteria and lower plants, which converts ribonucleotides to the DNA precursors, deoxyribonucleotides, a reaction in which a sugar -OH is replaced by -H. Note that ribonucleotide reductases catalyzing the same reaction in higher organisms and viruses are proteins with an oxo-bridged dimeric iron center.) The cobalt in vitamin B12 is coordinated to five N atoms, four contributed by a tetrapyrrole (corrin); the sixth ligand is C, provided either by C5 of deoxyadenosine in enzymes such as methylmalonyl-CoA mutase (fatty acid metabolism) or by a methyl group in the enzyme that synthesizes the amino acid methionine in bacteria.
Nickel is a component of a hydrolase (urease), of hydrogenase, of CO dehydrogenase, and of S-methyl CoM reductase, which catalyzes the terminal step in methane production by methanogenic bacteria. All the Ni-proteins known to date are from plants or bacteria.13,14 However, about 50 years elapsed between the crystallization of jack-bean urease in 1925 and the identification of the nickel component in the plant protein. Thus it is premature to exclude the possibility of Ni-proteins in animals. Despite the small number of characterized Ni-proteins, it is clear that many different environments exist, from apparently direct coordination to protein ligands (urease) to the tetrapyrrole F430 in methylreductase and the multiple metal sites of Ni and Fe-S in a hydrogenase from the bacterium Desulfovibrio gigas. Specific environments for nickel are also indicated for nucleic acids (or nucleic acid-binding proteins), since nickel activates the gene for hydrogenase.15
Manganese plays a critical role in oxygen evolution catalyzed by the proteins of the photosynthetic reaction center. The superoxide dismutase of bacteria and mitochondria, as well as pyruvate carboxylase in mammals, are also manganese proteins.16,17 How the multiple manganese atoms of the photosynthetic reaction center participate in the removal of four electrons and protons from water is the subject of intense investigation by spectroscopists, synthetic inorganic chemists, and molecular biologists.17
Vanadium and chromium have several features in common, from a bioinorganic viewpoint.18a First, both metals are present in only small amounts in most organisms. Second, the biological roles of each remain largely unknown.18 Finally, each has served as a probe to characterize the sites of other metals, such as iron and zinc. Vanadium is required for normal health, and could act in vivo either as a metal cation or as a phosphate analogue, depending on the oxidation state, V(lV) or V(V), respectively. Vanadium in a sea squirt (tunicate), a primitive vertebrate (Figure 1.2B), is concentrated in blood cells, apparently as the major cellular transition metal, but whether it participates in the transport of dioxygen (as iron and copper do) is not known. In proteins, vanadium is a cofactor in an algal bromoperoxidase and in certain prokaryotic nitrogenases. Chromium imbalance affects sugar metabolism and has been associated with the glucose tolerance factor in animals. But little is known about the structure of the factor or of any other specific chromium complexes from plants, animals, or bacteria.
Molybdenum proteins catalyze the reduction of nitrogen and nitrate, as well as the oxidation of aldehydes, purines, and sulfite.19 Few Mo-proteins are known compared to those involving other transition metals. Nitrogenases, which also contain iron, have been the focus of intense investigations by bioinorganic chemists and biologists; the iron is found in a cluster with molybdenum (the iron-molybdenum cofactor, or FeMoCo) and in an iron-sulfur center (Chapter 7). Interestingly, certain bacteria (Azotobacter) have alternative nitrogenases, which are produced when molybdenum is deficient and which contain vanadium and iron or only iron. All other known Mo-proteins are also Fe-proteins with iron centers, such as tetrapyrroles (heme and chlorins), Fe-sulfur clusters, and, apparently, non-heme/non-sulfur iron. Some Mo-proteins contain additional cofactors such as the Havins, e.g., in xanthine oxidase and aldehyde oxidase. The number of redox centers in some Mo-proteins exceeds the number of electrons transferred; reasons for this are unknown currently.

