14.19: \(^{13}C\) NMR Spectroscopy
- Page ID
- 16365
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)The 12C isotope of carbon - which accounts for up about 99% of the carbons in organic molecules - does not have a nuclear magnetic moment, and thus is NMR-inactive. Fortunately for organic chemists, however, the 13C isotope, which accounts for most of the remaining 1% of carbon atoms in nature, has a magnetic moment just like protons. Most of what we have learned about 1H-NMR spectroscopy also applies to 13C-NMR, although there are several important differences.
5.6A: The basics of 13C-NMR spectroscopy
The magnetic moment of a 13C nucleus is much weaker than that of a proton, meaning that NMR signals from 13C nuclei are inherently much weaker than proton signals. This, combined with the low natural abundance of 13C, means that it is much more difficult to observe carbon signals: more sample is required, and often the data from hundreds of scans must be averaged in order to bring the signal-to-noise ratio down to acceptable levels. Unlike 1H-NMR signals, the area under a 13C-NMR signal cannot be used to determine the number of carbons to which it corresponds. This is because the signals for some types of carbons are inherently weaker than for other types – peaks corresponding to carbonyl carbons, for example, are much smaller than those for methyl or methylene (CH2) peaks. Peak integration is generally not useful in 13C-NMR spectroscopy, except when investigating molecules that have been enriched with 13C isotope (see section 5.6B).
The resonance frequencies of 13C nuclei are lower than those of protons in the same applied field - in a 7.05 Tesla instrument, protons resonate at about 300 MHz, while carbons resonate at about 75 MHz. This is fortunate, as it allows us to look at 13C signals using a completely separate 'window' of radio frequencies. Just like in 1H-NMR, the standard used in 13C-NMR experiments to define the 0 ppm point is tetramethylsilane (TMS), although of course in 13C-NMR it is the signal from the four equivalent carbons in TMS that serves as the standard. Chemical shifts for 13C nuclei in organic molecules are spread out over a much wider range than for protons – up to 200 ppm for 13C compared to 12 ppm for protons (see Table 3 for a list of typical 13C-NMR chemical shifts). This is also fortunate, because it means that the signal from each carbon in a compound can almost always be seen as a distinct peak, without the overlapping that often plagues 1H-NMR spectra. The chemical shift of a 13C nucleus is influenced by essentially the same factors that influence a proton's chemical shift: bonds to electronegative atoms and diamagnetic anisotropy effects tend to shift signals downfield (higher resonance frequency). In addition, sp2 hybridization results in a large downfield shift. The 13C-NMR signals for carbonyl carbons are generally the furthest downfield (170-220 ppm), due to both sp2 hybridization and to the double bond to oxygen.
Because of the low natural abundance of 13C nuclei, it is very unlikely to find two 13C atoms near each other in the same molecule, and thus we do not see spin-spin coupling between neighboring carbons in a 13C-NMR spectrum. There is, however, heteronuclear coupling between 13C carbons and the hydrogens to which they are bound. Carbon-proton coupling constants are very large, on the order of 100 – 250 Hz. For clarity, chemists generally use a technique called broadband decoupling, which essentially 'turns off' C-H coupling, resulting in a spectrum in which all carbon signals are singlets. Below is the proton-decoupled13C-NMR spectrum of ethyl acetate, showing the expected four signals, one for each of the carbons.
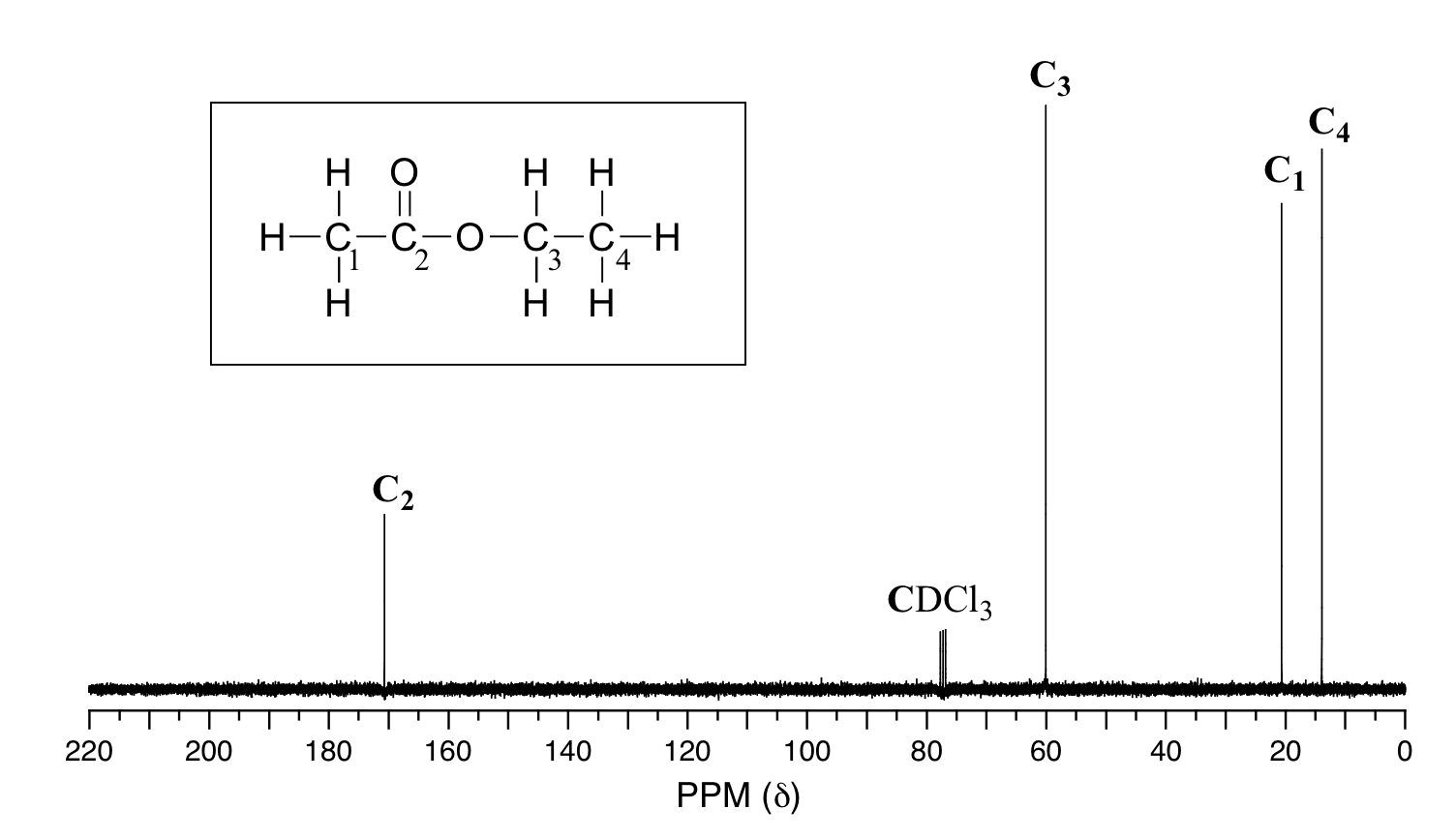
While broadband decoupling results in a much simpler spectrum, useful information about the presence of neighboring protons is lost. However, another modern NMR technique called DEPT (Distortionless Enhancement by Polarization Transfer) allows us to determine how many hydrogens are bound to each carbon. For example, a DEPT experiment tells us that the signal at 171 ppm in the ethyl acetate spectrum is a quaternary carbon (no hydrogens bound, in this case a carbonyl carbon), that the 61 ppm signal is from a methylene (CH2) carbon, and that the 21 ppm and 14 ppm signals are both methyl (CH3) carbons. The details of the DEPT experiment are beyond the scope of this text, but DEPT information will often be provided along with 13C spectral data in examples and problems.
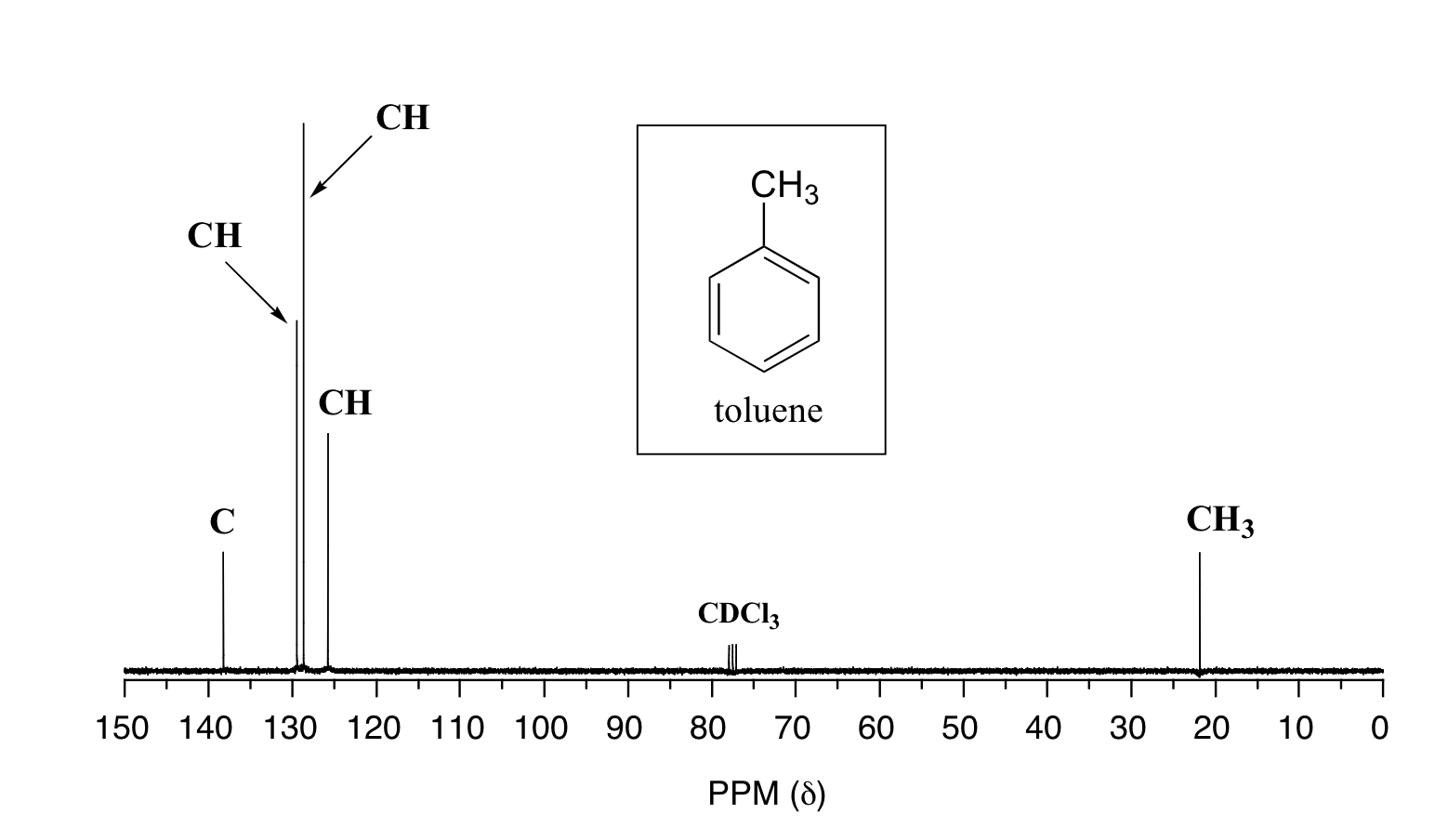
Below are two more examples of 13C NMR spectra of simple organic molecules, along with DEPT information.
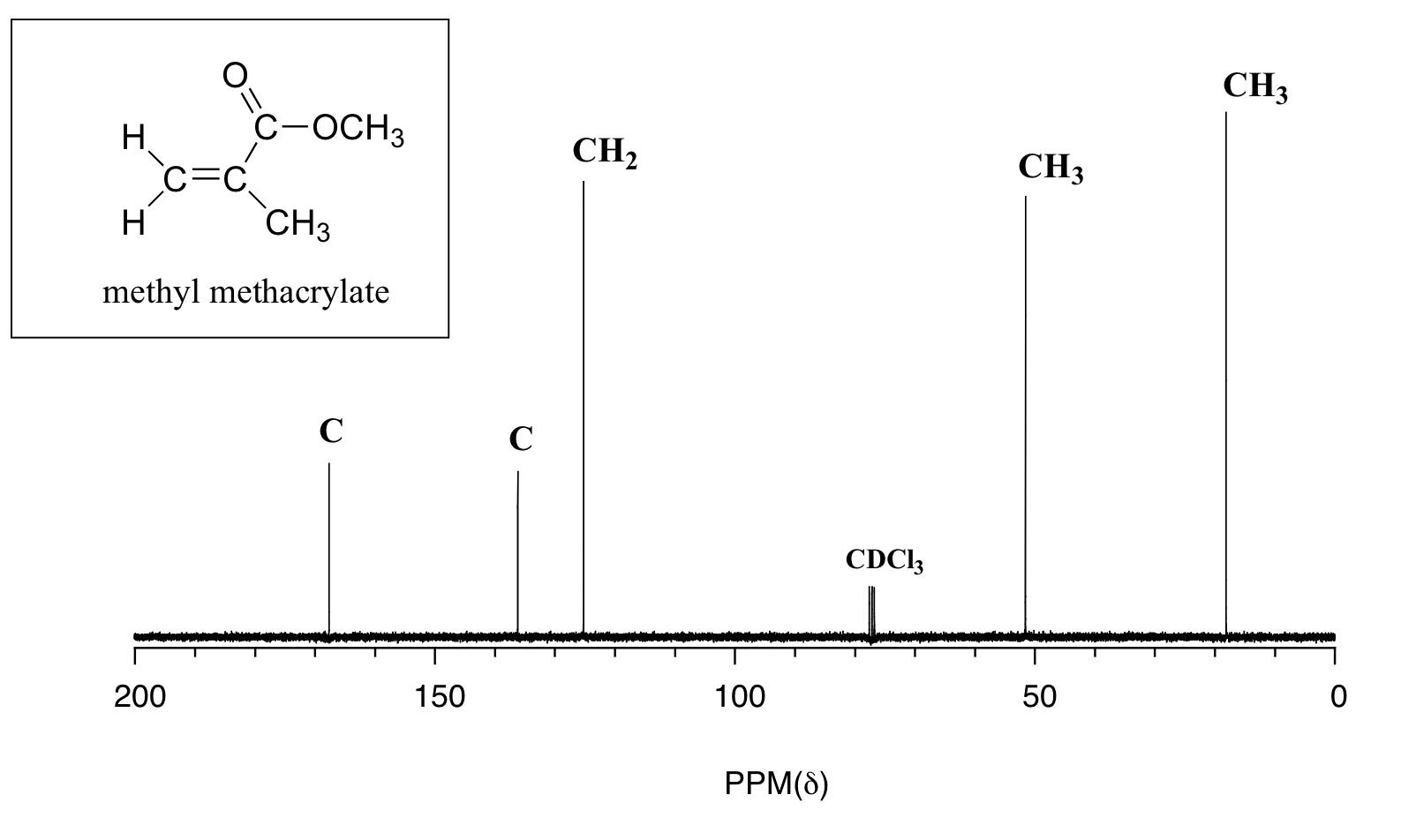
Example 5.13
Give peak assignments for the 13C-NMR spectrum of methyl methacrylate, shown above.
One of the greatest advantages of 13C-NMR compared to 1H-NMR is the breadth of the spectrum - recall that carbons resonate from 0-220 ppm relative to the TMS standard, as opposed to only 0-12 ppm for protons. Because of this, 13C signals rarely overlap, and we can almost always distinguish separate peaks for each carbon, even in a relatively large compound containing carbons in very similar environments. In the proton spectrum of 1-heptanol, for example, only the signals for the alcohol proton (Ha) and the two protons on the adjacent carbon (Hb) are easily analyzed. The other proton signals overlap, making analysis difficult.
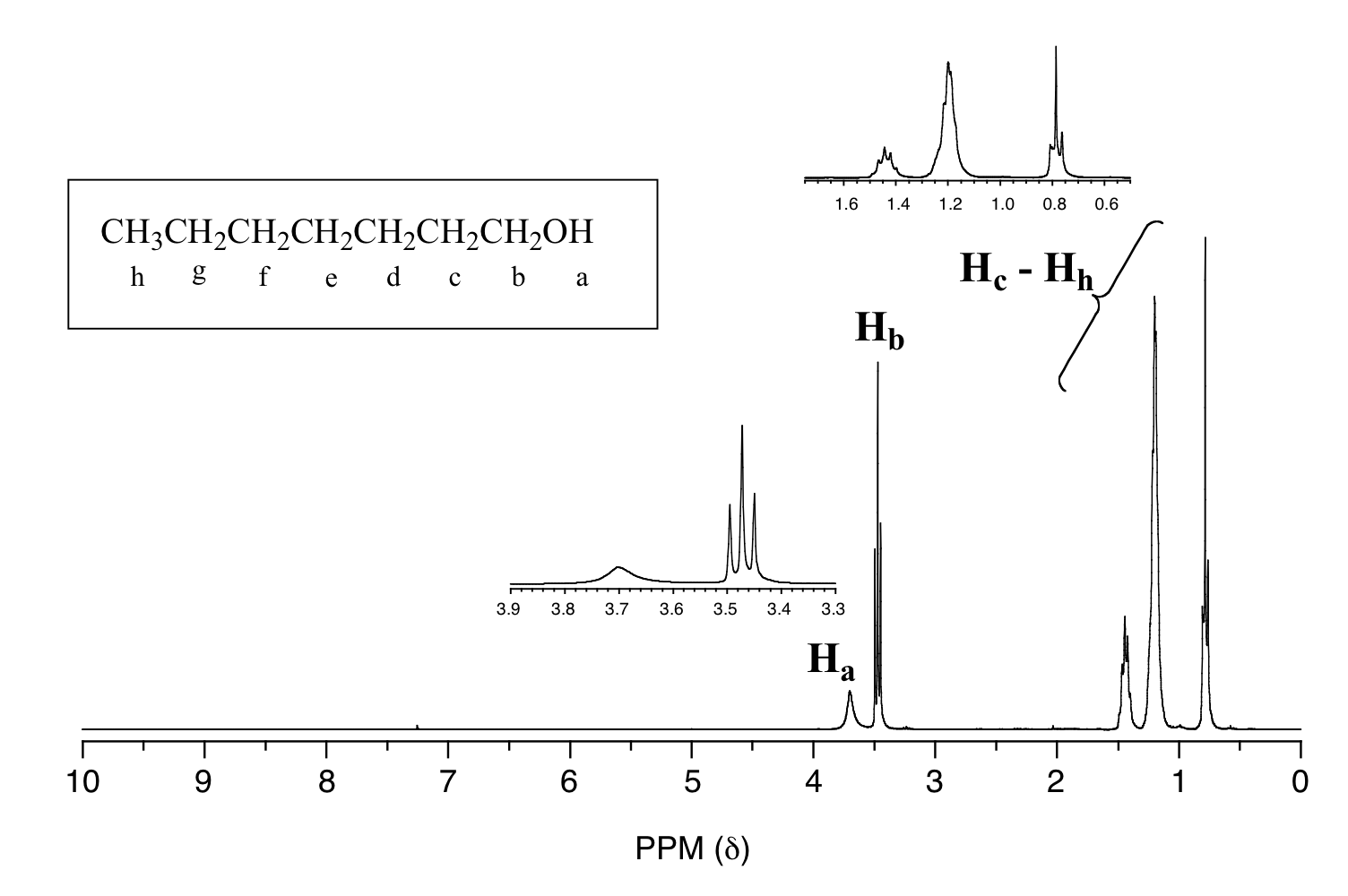
In the 13C spectrum of the same molecule, however, we can easily distinguish each carbon signal, and we know from this data that our sample has seven non-equivalent carbons. (Notice also that, as we would expect, the chemical shifts of the carbons get progressively smaller as they get farther away from the deshielding oxygen.)
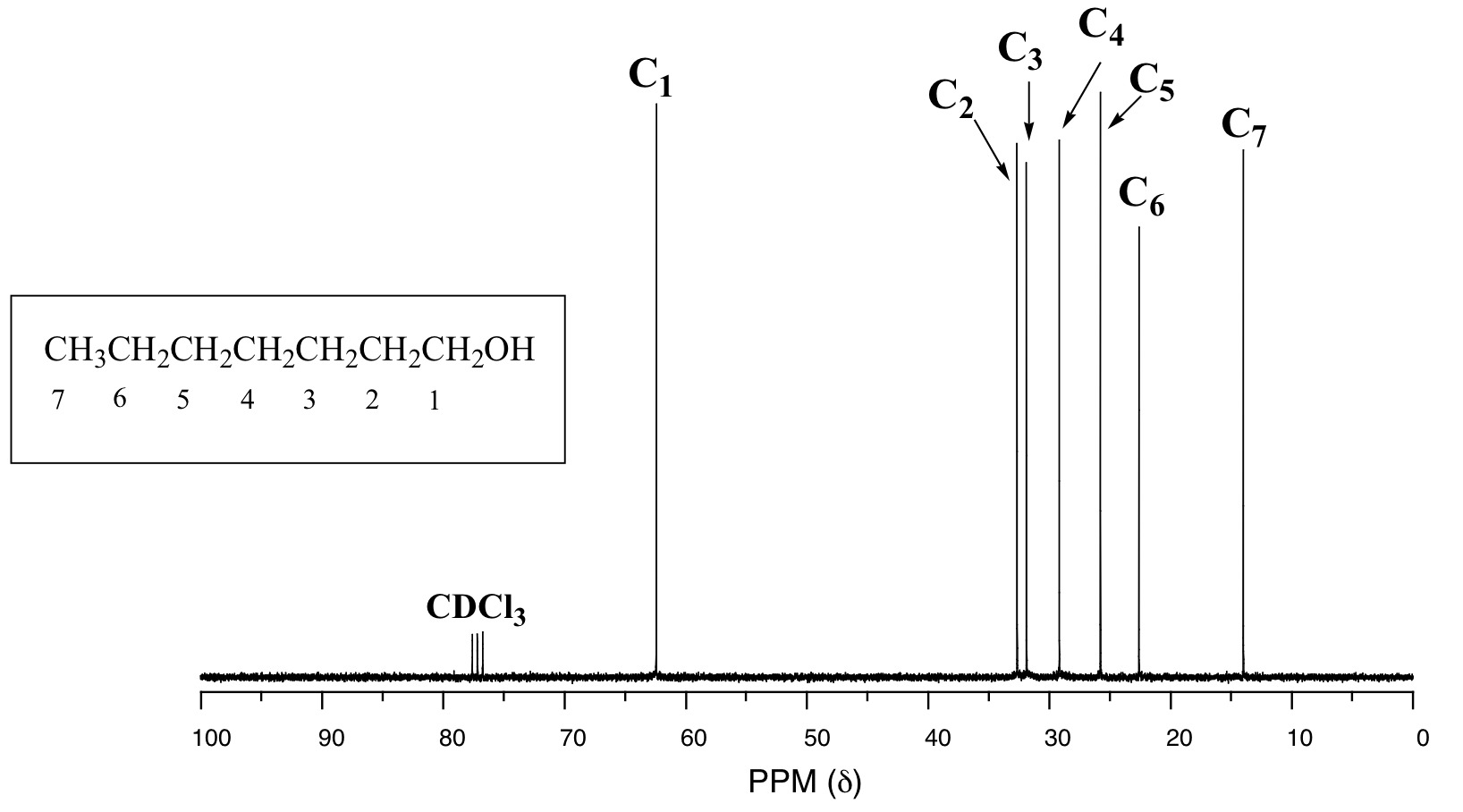
This property of 13C-NMR makes it very helpful in the elucidation of larger, more complex structures.
Example 5.14
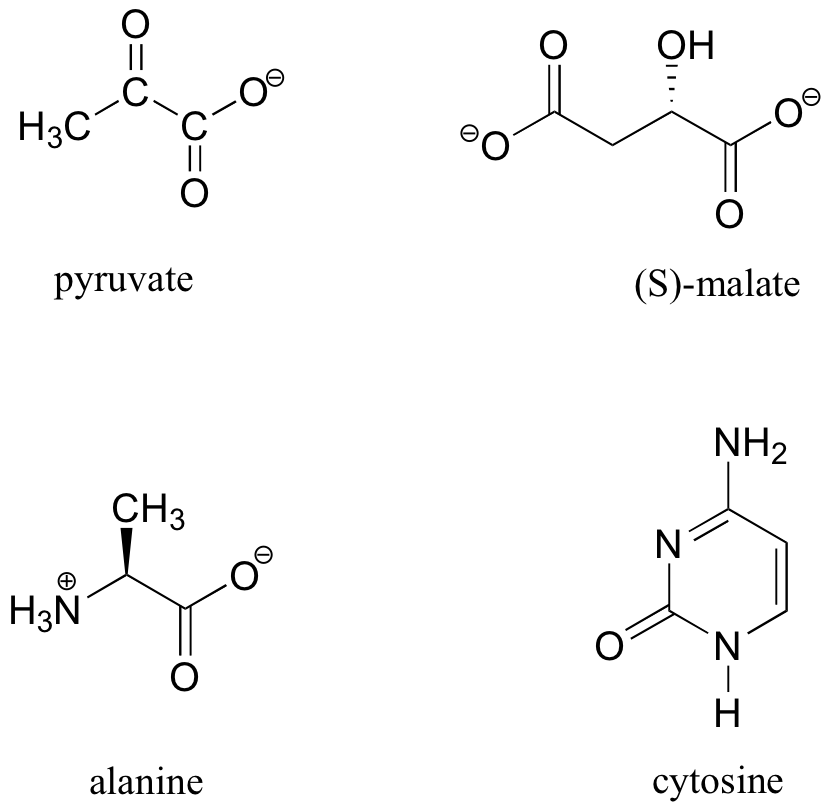
13C-NMR (and DEPT) data for some common biomolecules are shown below (data is from the Aldrich Library of 1H and 13C NMR). Match the NMR data to the correct structure, and make complete peak assignments.
- spectrum a: 168.10 ppm (C), 159.91 ppm (C), 144.05 ppm (CH), 95.79 ppm (CH)
- spectrum b: 207.85 ppm (C), 172.69 ppm (C), 29.29 ppm (CH3)
- spectrum c: 178.54 ppm (C), 53.25 ppm (CH), 18.95 ppm (CH3)
- spectrum d: 183.81 ppm (C), 182. 63 ppm (C), 73.06 ppm (CH), 45.35 ppm (CH2)
5.6B: \(^{13}C\)-NMR in isotopic labeling studies
Although only about 1 out of 100 carbon atoms in a naturally occurring organic molecule is the 13C isotope, chemists are often able to synthesize molecules that are artificially enriched in 13C at specific carbon positions, sometimes to the point where the 13C isotope is incorporated at a given position in over half of the molecules in the sample. This can be very useful, especially in biochemical studies, because it allows us to 'label' one or more carbons in a small precursor molecule and then trace the presence of the 13C label through a metabolic pathway all the way to a larger biomolecule product. For example, scientists were able to grow bacteria in a medium in which the only source of carbon was acetate enriched in 13C at the C1 (carbonyl) position. When they isolated a large molecule called amino-bacterio-hopanetriol (very similar in structure to cholesterol) from these bacteria and looked at its 13C-NMR spectrum, they observed that the 13C label from acetate had been incorporated at eight specific positions. They knew this because the 13C-NMR signals for these carbons were much stronger compared to the same signals in a control (unlabeled) molecule.
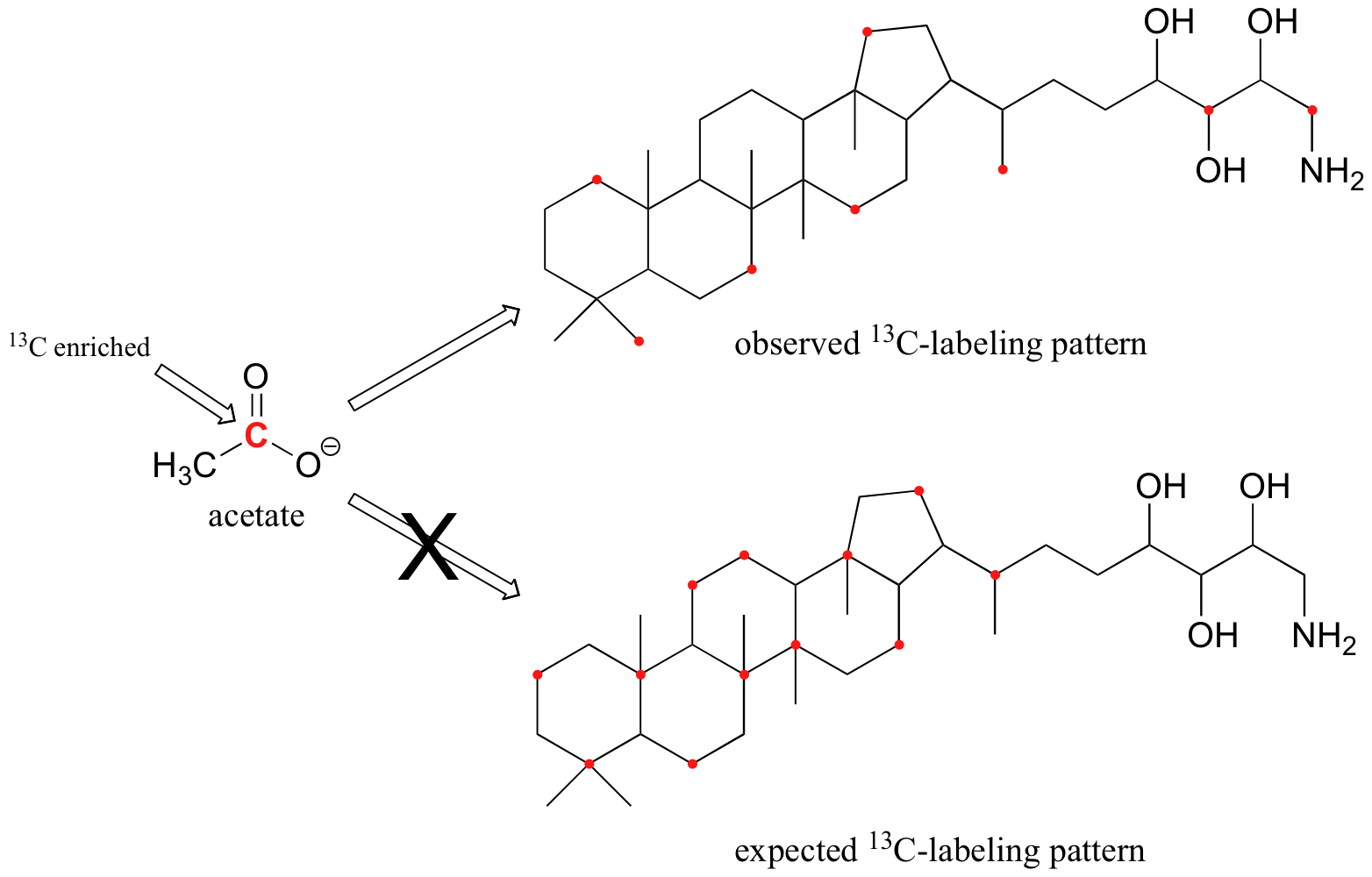
This result was very surprising - the scientists had expected a completely different pattern of 13C incorporation based on what they believed to be the metabolic pathway involved. This unexpected result led eventually to the discovery that bacteria synthesize these types of molecules in a very different way than yeasts, plants, and animals (Eur. J. Biochem. 1988, 175, 405). The newly discovered bacterial metabolic pathway is currently a key target for the development of new antibiotic and antimalaria drugs.

