Membrane Transport
- Page ID
- 2362
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Overview
Membrane transport is essential for cellular life. As cells proceed through their life cycle, a vast amount of exchange is necessary to maintain function. Transport may involve the incorporation of biological molecules and the discharge of waste products that are necessary for normal function.1
Membrane transport refers to the movement of particles (solute) across or through a membranous barrier.2 These membranous barriers, in the case of the cell for example, consist of a phospholipid bilayer. The phospholipids orient themselves in such a way so that the hydrophilic (polar) heads are nearest the extracellular and intracellular mediums, and the hydrophobic (non-polar) tails align between the two hydrophilic head groups.
Membrane transport is dependent upon the permeability of the membrane, transmembrane solute concentration, and the size and charge of the solute.2 Solute particles can traverse the membrane via three mechanisms: passive, facilitated, and active transport.1 Some of these transport mechanisms require the input of energy and use of a transmembrane protein, whereas other mechanisms do not incorporate secondary molecules.3
Passive transport
Passive transport is the simplest method of transport and is dependent upon the concentration gradient, and the size and charge of the solute.2 In passive transport, small uncharged solute particles diffuse across the membrane until both sides of the membrane have reached an equilibrium that is similar in concentration. The direction of solute travel is indicative of the concentration of that particular particle on each side of the membrane.
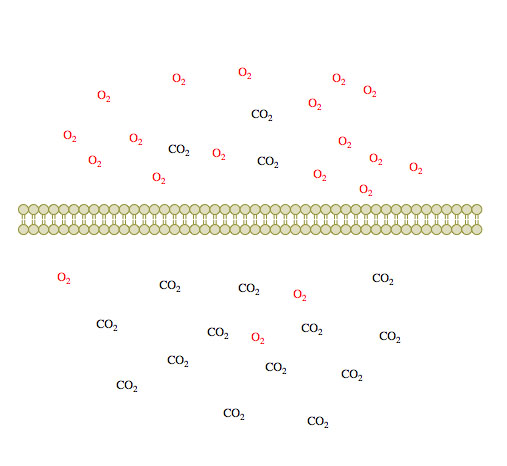 (1)
(1)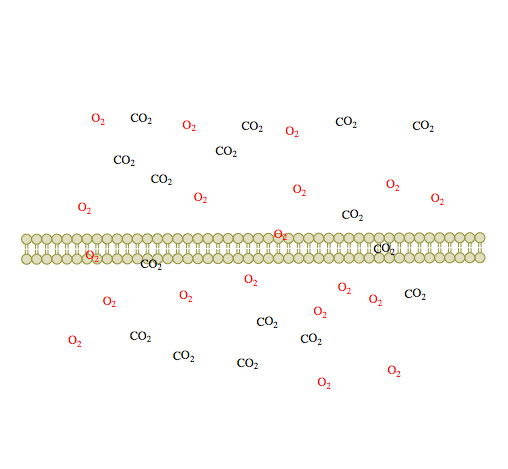 (2)
(2)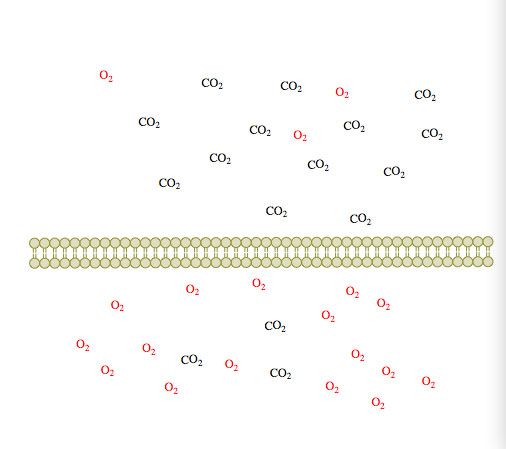 (3)
(3)
Figure 1. Passive diffusion of O2 and CO2 across a membrane over time 1-3.
Based on the thermodynamics of the system, particles will move from an area of high concentration to an area of low concentration in order to increase the entropy of the cell. Additionally, this particle movement will occur spontaneously as the free energy (Gibbs free energy; ∆G) of the system is negative.4
Where:
- Further, the amount of energy consumed or released by the system is as follows2:
- If ∆G<0, then particle movement is considered to be spontaneous; whereas, ∆G>0 particle movement requires the input of energy to move in the desired direction.
The properties of the membrane must also be considered when determining the rate of flow of the substrate.5 Darcy’s Law can be used to determine flow rate.
Where:
- The permeability coefficient is dependent upon the porous nature of the membrane and fluid.6
Where:
- Common permeability coefficients are displayed below7:
Membrane
Substrate
300
Escherichia coli
Glycerol
25
Escherichia coli
Glucose
1
Escherichia coli
Lactose
The diffusion of small charged particles, on the other hand, across a membrane is dependent upon the charge and transmembrane concentration of the solute.2 Again, however, the direction of solute travel is indicative of the thermodynamics of the system. Particles will travel from an area of high to low concentration, as well as travel so that the electrical potential across the membrane is diminished. As a result of this movement, the entropy of the system has increased.
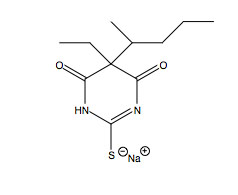
Passive transport is independent of membrane proteins and the catabolism of biological molecules for energy.2 This energy deficient process commonly occurs in the blood brain barrier as specific molecules, such as sodium thiopental (Figure 2. Sodium Thiopental Structure at right), can diffuse across the membrane.8 Sodium thiopental is a barbiturate frequently used in the methods of lethal injection.9 Sodium thiopental is a negatively charged particle and proceeds across the blood brain barrier to neuronal synaptic clefts. Sodium thiopental is an agonist of γ-aminobutyric acid (GABA), which acts as a neurotransmitter inhibitor. Sodium thiopental acts an anesthetic to cause unconsciousness.
Additionally, passive diffusion occurs across the placenta as all solute particles are exchanged between mother and fetus.10 Placental physiology juxtaposes maternal and fetal capillaries in order to exchange solute particles, such as oxygen and carbon dioxide gas.11 These uncharged molecules proceed across maternal and fetal capillary membranes in the direction from high to low concentration. This spontaneous process occurs in accord with the aforementioned properties. Despite the cellular system, passive diffusion across the membranes of all biological cells is in accord with the characteristics outlined above.
Facilitated Diffusion
Facilitated diffusion, not to be confused with simple diffusion, is a form of passive transport mediated by transport proteins imbedded within the cellular membrane.12 Facilitated diffusion allows the passage of lipophobic molecules through the cell membrane’s lipid bilayer.2 Just as in passive transport, molecules, particles, and ions travel freely across the cellular membrane from high concentration to low concentration in an attempt to achieve equilibrium and thereby increase the entropy of the system. Also like passive transport, the Gibbs Free Energy of the system is negative, allowing the particle movement to be spontaneous.4 Facilitated diffusion, however, uses channel proteins to facilitate solute movement.
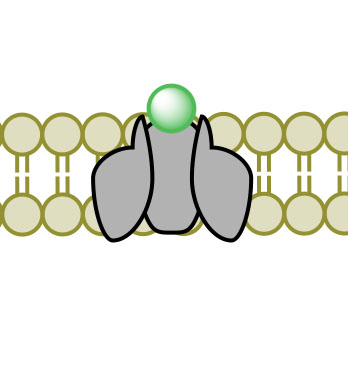 (1)
(1)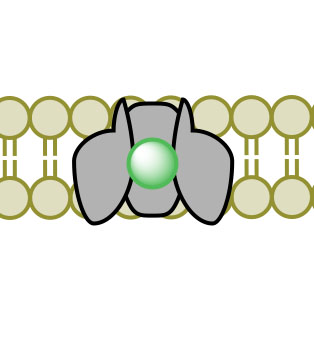 (2)
(2)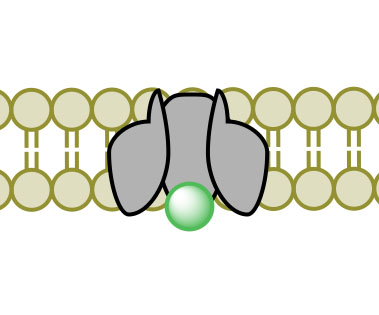 (3)
(3)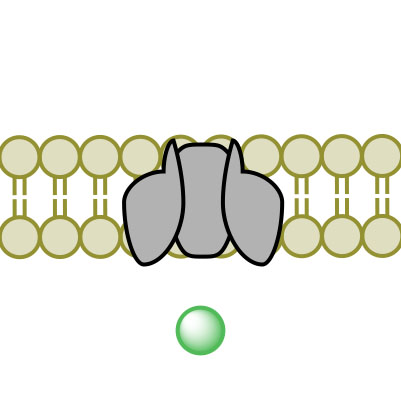 (4)
(4)Figure 3. Facilitated Diffusion via channel protein across a membrane proceeding images 1-4.
Channel Proteins
Channel proteins are pores immersed in the lipid bilayer membrane and are the hallmark of facilitated diffusion.13 All channel proteins have two things in common:
- It is of particular importance to note that channel proteins are not indiscriminate; each channel protein contains a selectivity filter.14 The selectivity filter is a collection of amino acid residues concentrated in the interior of the channel protein. As particles, often ions, pass into the channel protein, an electrostatic interaction occurs between the amino acid residues and the ion.15 The interaction would, for example, involve negatively charged amino acid residues in the case of ions like calcium (Na+) or potassium (K+), and positively charged amino acid residues in the case of chlorine (Cl-).2 The electrostatic interaction between amino acid residues and ions allows the channel protein to identify the ion in question by measuring its atomic radius with extremely finite accuracy. Potassium (K+) channels select K+ over Na+ by a factor of over one thousand despite differing in atomic radius by a mere 0.38 Å.
While all channel proteins have an inherent selectivity filter, others have additional gating.2 Gating is a response to a predetermined trigger that allows the channel protein to undergo a conformational change. This action subsequently causes another conformational change that either opens or closes the channel, allowing or disallowing its specific particle to pass. Channel proteins can be physically or chemically modulated through a number of different mechanisms.
Voltage gating
Voltage gated protein channels play a particularly important role in excitable neuronal and muscle tissues.2
Ligand gating
Ligand gated channel proteins are activated in response to the binding of a ligand.17 Typically, ligand binding occurs at an allosteric binding site independent of the channel protein’s pore. The binding of a ligand at the allosteric binding site causes a conformational change in the structure of the channel protein, subsequently causing an influx or efflux of ions. Release of the ligand allows the channel protein to return to its original shape. Structurally, ligand gated channel proteins generally differ from other channels due to the presence of an additional protein domain that serves as the allosteric binding site.2
The prototypical example of ligand gating is the nicotinic acetylcholine receptor located on the postsynaptic side of the neuromuscular junction.18
Other gating
Channel proteins may be gated in less common instances by methods such as light activation, mechanical activation, or secondary messanger activation.2Light activated protein channels contain a photoswitch through which a photon causes a conformational change in the channel protein causing it to open or close. Only one such protein channel exists naturally.19 Mechanically activated protein channels open or close in response to a mechanical stimulus and are vital to the touch, hearing, and balance sensations in human.20 Ligand-gated protein channels are typically linked to second messanger gating.2 Second messenger gating functions stepwise in that a neurotransmitter binds to a channel protein receptor which, in turn, reveals an active site to which the conformation-changing ligand binds.
Active Transport
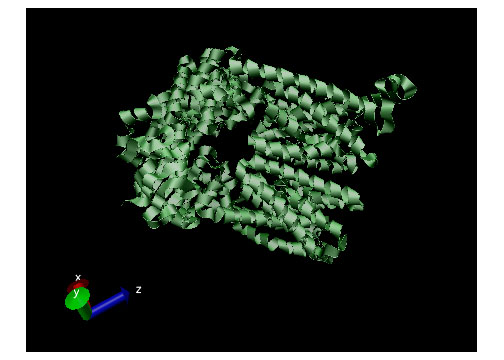 Active transport, simply put, is the movement of particles through a transport protein from low concentration to high concentration at the expense of metabolic energy.21 The most common energy source used by cells is adenosine triphosphate or ATP, though other sources such as light energy or the energy stored in an electrochemical gradient are also utilized.2 In the case of ATP, energy is chemically harvested through hydrolysis.22 ATP hydrolysis in turn causes a conformational change in the transport protein which allows mechanical movement of the particle in question.2 Active transport systems are, therefore, energy-coupling devices as chemical and mechanical processes are linked to achieve particle movement. Active transport is classified as either Primary Active Transport or Secondary Active Transport. Figure 4 (displayed above) displays a ribbon structure of a commonly depicted ABC Vitamin B12importer active transport protein.
Active transport, simply put, is the movement of particles through a transport protein from low concentration to high concentration at the expense of metabolic energy.21 The most common energy source used by cells is adenosine triphosphate or ATP, though other sources such as light energy or the energy stored in an electrochemical gradient are also utilized.2 In the case of ATP, energy is chemically harvested through hydrolysis.22 ATP hydrolysis in turn causes a conformational change in the transport protein which allows mechanical movement of the particle in question.2 Active transport systems are, therefore, energy-coupling devices as chemical and mechanical processes are linked to achieve particle movement. Active transport is classified as either Primary Active Transport or Secondary Active Transport. Figure 4 (displayed above) displays a ribbon structure of a commonly depicted ABC Vitamin B12importer active transport protein.Primary Active Transport
Primary active transport uses the energy found in ATP, photons, and electrochemical gradients directly in the transport of molecules from low concentration to high concentration across the cellular membrane.23
Using ATP
The enzyme-catalyzed hydrolysis reaction removing a phosphate from ATP, thereby forming ADP, causes a conformational change in the transport protein allowing particles to influx or efflux.23 Enzymes catalyzing ATP-driven primary active transport are called ATPases.2
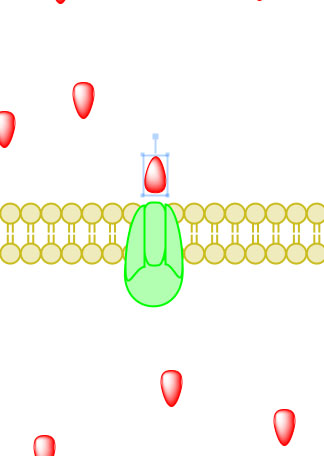
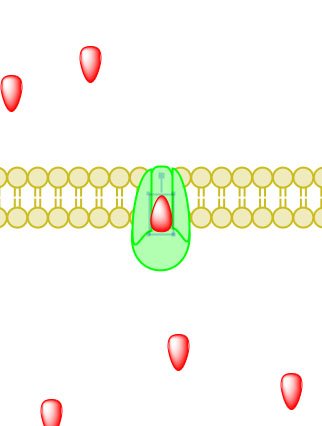
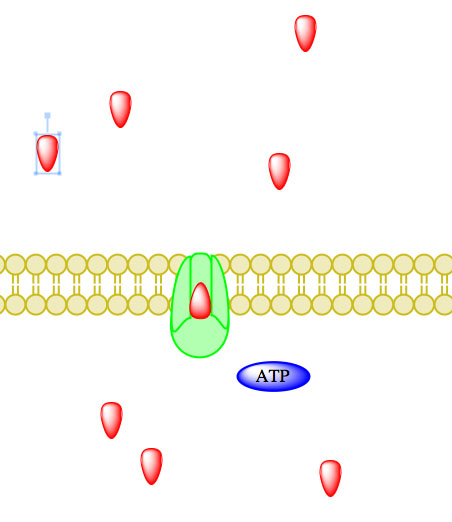
_____________________________________________________________________________________
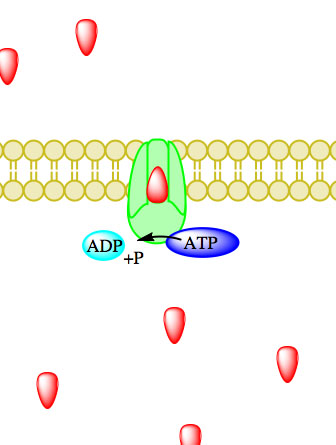
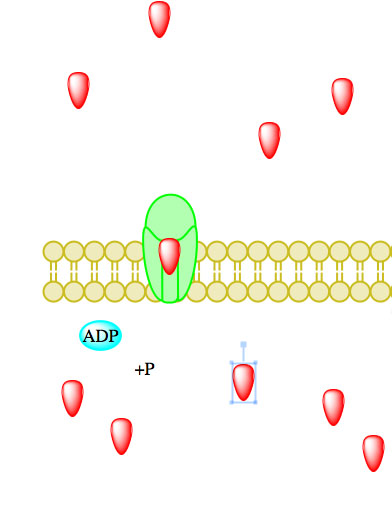
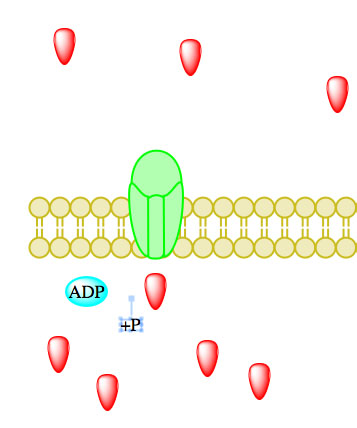
Figure 5. Primary active transport, with the use of ATP, is depicted above progressing left to right and top to bottom.
The most universal example of ATP hydrolysis driving primary active transport in cells is the sodium-potassium pump.2 The sodium-potassium pump is responsible for controlling both sodium and potassium concentrations inside the cell. The sodium-potassium pump is extremely important in maintaining the cell’s resting potential.
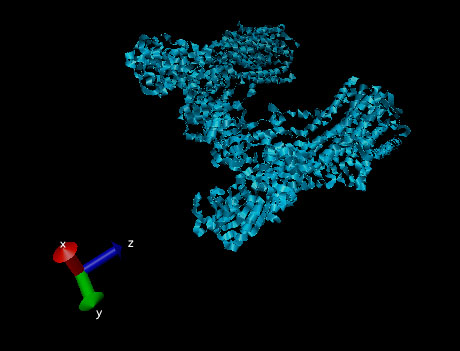
Figure 6. Ribbon structure of a sodium-potassium ATPase pump.
Using Electrochemical Gradient Energy
An electrochemical gradient has two components: 1) an electrical component caused by charge difference on either side of the cellular membrane and 2) a chemical component resulting from differing concentrations of ions across the cellular membrane.24 The electrochemical gradient is generated by the presence of a proton (H+) gradient. A proton gradient is an interconvertible form of energy that can ultimately be used by the transport protein to move particles across the cellular membrane.2
A quintessential example of electrochemical gradient energy in primary active transport is the mitochondrial electron transport chain (ETC).25 The ETC uses the energy produced from the reduction of NADH to NAD+ to create a proton gradient by pumping protons into the inner mitochondrial space.
Using Photon Energy
The energy stored in a photon, the basic unit of light, is used to generate a proton gradient through a process similar to that found in electrochemical gradients.24 The stepwise passing of electrons in an electron transport chain reduces a molecule like NADH and ultimately generates a proton gradient.
Plant photosynthesis is an example of primary active transport using photon energy.2 Chlorophyll absorbs a photon of light and consequently loses an electron which it passes pheophytin causing a subsequent electron transport chain.26 This ETC ultimately ends in the reduction of NADH to NAD+ creating a proton gradient across the chloroplast membrane.
Secondary Active Transport
Secondary active transport achieves an identical result as primary active transport in that particles are moved from low concentration to high concentration at the expense of energy.2 Secondary active transport, however, functions independent of direct ATP coupling. Rather, the electrochemical energy generated from pumping ions out of the cell is used. Secondary active transport is classified as either symporter of antiporter.
Symports
Symport secondary active transport uses a downhill movement of one particle to transport another particle against its concentration gradient.27 Symports move both particles in the same direction through a transmembrane transport protein.
A common symport example is SGLT1, a glucose symport. SGLT1 tranports one glucose molecule into the cell for every two sodium ions transported into the cell.28 The SGLT1 symport is located throughout the body, particularly in the nephron of the kidney.
Antiports
Antiport secondary active transport moves two or more different particles across the cellular membrane in opposite directions.27 Antiport secondary active transport moves one particle down its concentration gradient and uses the energy generated from that process to move another particle up its concentration gradient.
The sodium-calcium exchanger found throughout humans in excitable cells is a simple and common example of an antiport. Three sodium ions travel down their concentration gradient in exchange for one calcium ion.29
References
(1a, b) Pardee, A. Membrane transport proteins: proteins that appear to be parts of membrane transport systems are being isolated and characterized. Science, 1968, 162, 632-637. DOI: 10.1126/science.162.3854.632
(2a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t, u) Garrett, R .; Grisham, C. Biochemistry, 4th ed.; Brooks/Cole: Boston, 2009.
(3^) Stein, W. D. The Movement of Molecules Across Cell Membranes; Stein, W. D., Eds, Academic Press: New York, 1967.
(4a, b) Alberty, R. Standard Gibbs free energy, enthalpy, and entropy changes as a function of pH and pMg for several reactions involving adenosine phosphates. The Journal of Biological Chemistry, 1969, 244, 3290-3302. PMID: 4307313
(5^) Pal, L.; Joyce, M.; Fleming, P. A simple method for calculation of the permeability coefficient of porous media. Peer-Reviewed Paper Physics: TAPPI Journal, 2006, 5, 10-16.
(6^) Walter, A.; Gutknecht, J. Permeability of small nonelectrolytes through lipid bilayer membranes. Journal of Membrane Biology, 1986, 90, 207-217. DOI:10.1007/BF01870127
(7^) Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiological Reviews, 1985, 49, 1-32. PMCID: PMC373015
(8^) Fischer, H.; Gottschlich, R.; Seelig, A. Blood-brain barrier permeation: molecular parameters governing passive diffusion. J. Membrane Biol.,1998, 165, 201-211. DOI: 10.1007/s002329900434
(9^) Jevtovic-Todorovic, V.; Wozniak, D.; Powell, S.; Olney, J. Propofol and sodium thiopental protect against MK-801-induced neuronal necrosis in the posterior cingulated/retrosplenial cortex. Brian Research, 2001, 913, 185-189. PMID: 11549385
(10^) Sibley, C.; Boyd, R. Control of transfer across the mature placenta. Oxf. Rev. Reprod. Biol., 1988, 10, 382-435. PMID: 3072506
(11^) Reynolds, L.; Redmer, D. Utero-placental vascular development and placental function. Journal of Animal Science, 1995, 73, 1839-1851. PMID: 7545661
(12^) Cusslera, E.; Arisa, R.; Bhown, A. On the limits of facilitated diffusion. Journal of Membrane Science, 1989, 2-3, 149-164. DOI:10.1016/S0376-7388(00)85094-2
(13^) Hille, B. Ion channels of excitable membranes, 3rd ed. Sinauer Associates: Sunderland, MA, 2001.
(14^) Hellgren M.; Sandberg L.; Edholm O. A comparison between two prokaryotic potassium channels (KirBac1.1 and KcsA) in a molecular dynamics (MD) simulation study. Biophysical Chemistry, 2006,120, 1–9. DOI: 10.1016/j.bpc.2005.10.002
(15^) Coulomb, C-A. Recherches théoriques et expérimentales sur la force de torsion et sur l'élasticité des fils de metal. Histoire de l’Académie Royale des Sciences, 1784, 229-269.
(16^) Catterall, W. Structure and function of voltage-gated ion channels. Annual Review of Biochemistry, 1995, 64, 493-531. DOI: 10.1016/0166-2236(93)90193-P
(17^) Sheng, M.; Pak, D. Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annual Review of Physiology, 2000, 62, 755-78. DOI:10.1146/annurev.physiol. 62.1.755
(18^) Purves, D.; Augustine, G.; Fitzpatrick, D.; Hall, W.; LaMantia, A.; McNamara, J.; White, L. Neuroscience, 4th ed. Sinauer Associates: Sunderland, MA, 2008.
(19^) Banghart, M.; Volgraf, M.; Trauner, D. Engineering light-gated ion channels. American Chemical Society, 2006, 45, 15129–15141. DOI: 10.1021/bi0618058
(20^) Pivetti, C.; Yen, M.; Miller, S.; Busch, W.; Tseng, Y.; Booth, I.; Saier, M. Two Families of Mechanosensitive Channel Proteins. Microbiology and Molecular Biology Reviews, 2003, 67, 66-85. DOI: 10.1128/MMBR.67.1.66-85.2003
(21^) Hodgkin, A.; Keynes, R. Active transport of cations in giant axons from sepia and loligo. The Journal of Physiology, 1955, 128, 28-60. PMC1365754
(22^) Nicholls, D. The influence of respiration and ATP hydrolysis on the proton-electrochemical gradient across the inner membrane of rat-liver mitochondria as determined by ion distribution. European Journal of Biochemistry, 1974, 50, 305-315. DOI: 10.1111/j.1432-1033.1974. tb03899.x
(23a, b) Mandel, L. Primary active sodium transport, oxygen consumption, and ATP: Coupling and regulation. Kidney International, 1986, 29, 3-9. DOI: 10.1038/ki.1986.2
(24a, b) Zilberstein, D.; Schuldiner, S.; Padan, E. Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry, 1979, 18, 669-673. DOI: 10.1021/bi00571a018
(25^) Karp, G. Cell and Molecular Biology,5th ed. John Wiley & Sons: Hoboken, NJ, 2008.
(26^) Raven, P.; Evert, R.; Eichhorn, S. Biology of Plants, 7th ed. W.H. Freeman and Company Publishers: New York, 2005.
(27a, b) Pâun, A.; Pâun, G. The power of communication: P systems with symport/antiport. New Generation Computing, 2002, 20, 295-305. DOI: 10.1007/BF03037362
(28^) Wright, E.; Hirayama B.; Loo D. Active sugar transport in health and disease. Journal of Internal Medicine, 2007, 261, 32–43. DOI: 10.1111/j.1365-2796.2006.01746.x
(29^) Yu, S.; Choi, D. Na+–Ca2+ exchange currents in cortical neurons: concomitant forward and reverse operation and effect of glutamate. European Journal of Neuroscience, 1997, 1273–81. PMID: 9215711
Contributors:
- Garret Powell (Truman State University)
- Jordan Kaminski (Truman State University)
- It is of particular importance to note that channel proteins are not indiscriminate; each channel protein contains a selectivity filter.14 The selectivity filter is a collection of amino acid residues concentrated in the interior of the channel protein. As particles, often ions, pass into the channel protein, an electrostatic interaction occurs between the amino acid residues and the ion.15 The interaction would, for example, involve negatively charged amino acid residues in the case of ions like calcium (Na+) or potassium (K+), and positively charged amino acid residues in the case of chlorine (Cl-).2 The electrostatic interaction between amino acid residues and ions allows the channel protein to identify the ion in question by measuring its atomic radius with extremely finite accuracy. Potassium (K+) channels select K+ over Na+ by a factor of over one thousand despite differing in atomic radius by a mere 0.38 Å.
- Common permeability coefficients are displayed below7:
- The permeability coefficient is dependent upon the porous nature of the membrane and fluid.6
- If ∆G<0, then particle movement is considered to be spontaneous; whereas, ∆G>0 particle movement requires the input of energy to move in the desired direction.

