Nitrogenase 1
- Page ID
- 2507
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\) There are numerous types of enzymes, complexes, and o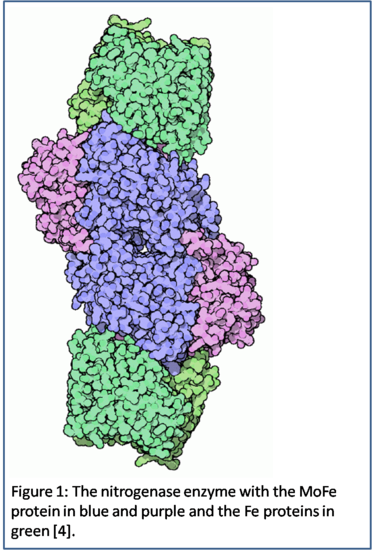 ther material that operate in living organisms and can be important to their survival. Nitrogenase, shown in Figure 1, is one enzyme that is produced by certain types of bacteria and is vital to their existence and growth. Nitrogenase is a unique enzyme with a crucial function that is distinct to bacteria that utilize it, has unique structure and symmetry, and is sensitive to other compounds that inhibits its functioning. It is “an enzymatic complex which enables fixation of atmospheric nitrogen” [3]. The unique structure of nitrogenase is almost completely known because of the extensive research that has been done on this enzyme. Nitrogenase can also bind to compounds other than nitrogen gas, which can inhibit and decrease its production of ammonia to the rest of the organism’s body. Without proper functioning, the bacteria that utilize nitrogenase would not be able to survive, and other organisms that depend on these bacteria would also die.
ther material that operate in living organisms and can be important to their survival. Nitrogenase, shown in Figure 1, is one enzyme that is produced by certain types of bacteria and is vital to their existence and growth. Nitrogenase is a unique enzyme with a crucial function that is distinct to bacteria that utilize it, has unique structure and symmetry, and is sensitive to other compounds that inhibits its functioning. It is “an enzymatic complex which enables fixation of atmospheric nitrogen” [3]. The unique structure of nitrogenase is almost completely known because of the extensive research that has been done on this enzyme. Nitrogenase can also bind to compounds other than nitrogen gas, which can inhibit and decrease its production of ammonia to the rest of the organism’s body. Without proper functioning, the bacteria that utilize nitrogenase would not be able to survive, and other organisms that depend on these bacteria would also die.
Nitrogenase is unique in its ability to fix nitrogen, so that it is more reactive and able to be applied in other reactions that help organisms grow and thrive. David Goodsell states, “Nitrogen is needed by all living things to build proteins and nucleic acids” [4]. However, nitrogen gas, N2, is an inert gas that is stabilized by its triple bond [5], and is difficult for living organisms to use as a source of nitrogen because the molecule’s stability. Nitrogenase is used to separate nitrogen gas, N2, and transforms it into ammonia, NH3 in the reaction:
N2 + 8H+ + 8e- + 16 ATP + 16H2O ----> 2NH3 + H2 + 16ADP + 16Pi
in room temperature and atmospheric pressure [1].
In the form of ammonia organisms have a useable source of nitrogen that is more reactive and can be used to 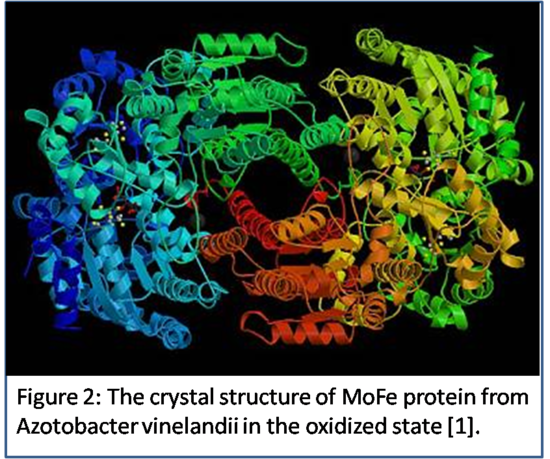 create proteins and nucleic acids that are also necessary for the organism. According to the Peters and Szilagyi, “Three types of nitrogenase are known, called molybdenum (Mo) nitrogenase, vanadium (V) nitrogenase and iron-only (Fe) nitrogenase” and the molybdenum nitrogenase,crystal structure shown in Figure 2, is the one that has been studied the most of the three [6]. The nitrogenase enzyme breaks up a diatomic nitrogen gas molecule using a large number of ATP and 8 electrons to create two ammonia molecules and hydrogen gas for each molecule of nitrogen gas [4]. As a result, the bacteria that utilize this enzyme must expend much of their energy, in the form of ATP, so that they will constantly obtain a steady source of nitrogen. Without nitrogenase’s function of fixing nitrogen gas into ammonia, then organisms would not be able to thrive since they would not receive a source of nitrogen for other important reactions.
create proteins and nucleic acids that are also necessary for the organism. According to the Peters and Szilagyi, “Three types of nitrogenase are known, called molybdenum (Mo) nitrogenase, vanadium (V) nitrogenase and iron-only (Fe) nitrogenase” and the molybdenum nitrogenase,crystal structure shown in Figure 2, is the one that has been studied the most of the three [6]. The nitrogenase enzyme breaks up a diatomic nitrogen gas molecule using a large number of ATP and 8 electrons to create two ammonia molecules and hydrogen gas for each molecule of nitrogen gas [4]. As a result, the bacteria that utilize this enzyme must expend much of their energy, in the form of ATP, so that they will constantly obtain a steady source of nitrogen. Without nitrogenase’s function of fixing nitrogen gas into ammonia, then organisms would not be able to thrive since they would not receive a source of nitrogen for other important reactions.
Many details have been discovered over time about the functioning of nitrogenase, but there has yet to a complete agreement on the structure of nitrogenase. The iron-molybdenum cofactor center of the nitrogenase 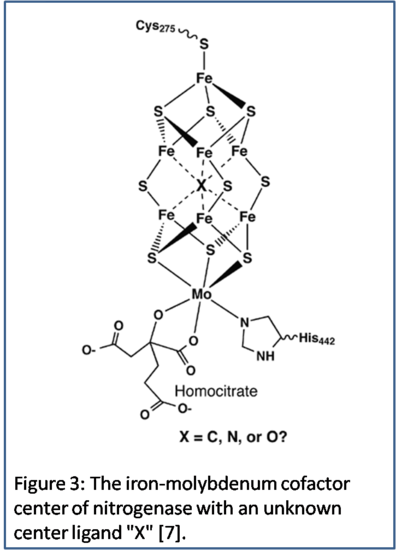 enzyme consists of iron, sulfur, molybdenum, a homocitrate molecule, a histadine amino acid and a cysteine amino acid [7]. There also exists the possibility of more to the structure that is not yet known to scientists, such as the possibility of a central atom within the cofactor center of nitrogenase, shown in Figure 3 with possible C, N or O as a central atom [7]. Nitrogenase contains various molecules that have different conformations throughout the enzyme giving nitrogenase a unique symmetry. The metals in the iron-molybdenum cofactor center of nitrogenase have three different geometrical conformations and a unique symmetry to each of these metals as a result. The lone iron that is closest to cysteine has a tetrahedral conformation, and a Td’ symmetry as a result. The six irons near the center have a trigonal planar conformation with D3h symmetry, and possibly a different conformation and symmetry if a central core atom is discovered. Lastly, the one molybdenum atom has an octahedral conformation with a C1 symmetry because of the asymmetric homocitrate that it is connected to. Overall, the iron-molybdenum cofactor center of nitrogenase has a C1 symmetry, which is mostly due to the asymmetric connections that molybdenum has to homocitrate and histadine [7]. This is possibly the reason why there are not many IR and Raman spectroscopes that provide useful information to the structure of nitrogenase because of its lack of symmetry. However, other forms of spectroscopy, such as X-ray, provide more information about the iron atoms found within nitrogenase [8]. Nitrogenase has a unique structure and as science continues to advance the finalized structure will eventually be agreed upon by scientists.
enzyme consists of iron, sulfur, molybdenum, a homocitrate molecule, a histadine amino acid and a cysteine amino acid [7]. There also exists the possibility of more to the structure that is not yet known to scientists, such as the possibility of a central atom within the cofactor center of nitrogenase, shown in Figure 3 with possible C, N or O as a central atom [7]. Nitrogenase contains various molecules that have different conformations throughout the enzyme giving nitrogenase a unique symmetry. The metals in the iron-molybdenum cofactor center of nitrogenase have three different geometrical conformations and a unique symmetry to each of these metals as a result. The lone iron that is closest to cysteine has a tetrahedral conformation, and a Td’ symmetry as a result. The six irons near the center have a trigonal planar conformation with D3h symmetry, and possibly a different conformation and symmetry if a central core atom is discovered. Lastly, the one molybdenum atom has an octahedral conformation with a C1 symmetry because of the asymmetric homocitrate that it is connected to. Overall, the iron-molybdenum cofactor center of nitrogenase has a C1 symmetry, which is mostly due to the asymmetric connections that molybdenum has to homocitrate and histadine [7]. This is possibly the reason why there are not many IR and Raman spectroscopes that provide useful information to the structure of nitrogenase because of its lack of symmetry. However, other forms of spectroscopy, such as X-ray, provide more information about the iron atoms found within nitrogenase [8]. Nitrogenase has a unique structure and as science continues to advance the finalized structure will eventually be agreed upon by scientists.
Nitrogenase is not easily used because of its sensitivity to other compounds, which prevents it from being easily used by other organisms. According to Diskin, “The nitrogenase complex is very sensitive to oxygen,” [3] and as a result all aerobic organisms are unable to utilize this enzyme. Nitrogenase contains several 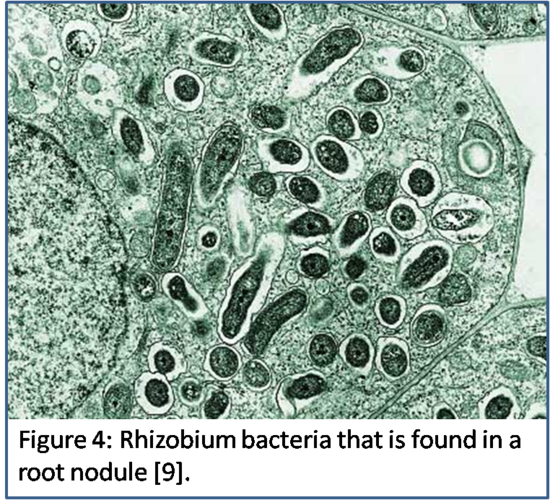 iron atoms that could become disrupted if it were to become oxidized by any oxygen gas. In addition, “nitrogenase has the ability to bond acetylene and carbon monoxide” [10] making the three molecules (acetylene, carbon monoxide, and nitrogen gas) competitive substrates that bind to nitrogenase. The restrictive functionality of nitrogenase makes it only possible for anaerobic organisms to utilize nitrogenase. Rhizobium (Figure 4), normally found in plant roots, is an example of an anaerobic family of bacteria that utilizes nitrogenase. They exist in a symbiotic relationship with leguminous plants to produce nitrogen in exchange for other nutrients and protection from oxygen and other compounds that could inhibit nitrogenase’s operation [2]. These limitations of nitrogenase prevent it from being commonly used among organisms, but it is an important element to the bacteria that make use of this enzyme.
iron atoms that could become disrupted if it were to become oxidized by any oxygen gas. In addition, “nitrogenase has the ability to bond acetylene and carbon monoxide” [10] making the three molecules (acetylene, carbon monoxide, and nitrogen gas) competitive substrates that bind to nitrogenase. The restrictive functionality of nitrogenase makes it only possible for anaerobic organisms to utilize nitrogenase. Rhizobium (Figure 4), normally found in plant roots, is an example of an anaerobic family of bacteria that utilizes nitrogenase. They exist in a symbiotic relationship with leguminous plants to produce nitrogen in exchange for other nutrients and protection from oxygen and other compounds that could inhibit nitrogenase’s operation [2]. These limitations of nitrogenase prevent it from being commonly used among organisms, but it is an important element to the bacteria that make use of this enzyme.
Nitrogenase is a unique enzyme that is critical to the bacteria that utilizes this enzyme. Its function is necessary for anaerobic bacteria since it is their main source of nitrogen. However, the high cost of ATP and electrons is another restrictive reason why more organisms do not produce their own source of nitrogen. The specific connections within nitrogenase cause it to lack any symmetric elements in its structure. More research must be done before the complete understanding of nitrogenase and its structure, such as with the possibility and identity to a central atom within nitrogenase. Nitrogenase is a sensitive enzyme that can easily be inhibited by oxygen and other compounds and affects its ability to fix nitrogen. It is the major reason why nitrogenase is strictly found in anaerobic bacteria that is able to utilize nitrogenase for its source of nitrogen. Nitrogenase is simply one enzyme among the multitudes of other enzymes and complexes that is vital strictly to the organisms that utilize it.
References
1. Achim,Catalina. “Magneto-electronics of Transition-metal Clusters.” The Achim Lab. Achim,Catalina. 22 May 2010. <http://www.chem.cmu.edu/groups/achim...h/magneto.html>.
2. Clark, David P. “Nitrogen Fixation.” Microbiology 425: Physiology & Biochemistry of Microorganisms. 3 Aug 99. 23 May 2010. <http://www.micro.siu.edu/Micr425/425...2-NitrFix.html>.
3. Diskin, Tali. “The Nitrogenase Complex.” Nif genes. 22 May 2010. <http://www.tau.ac.il/~ecology/virtau...%20complex.htm>.
4. Goodsell, David S. “Nitrogenase.” RCSB Protein Data Bank. Feb 2002. 22 May 2010. <http://www.rcsb.org/pdb/static.do?p=...h/pdb26_1.html>.
5. Johnson, Erik. “Physical Properties of Nitrogen Gas.” Ehow. 23 May 2010. <http://www.ehow.com/about_5382428_ph...rogen-gas.html>.
6. “Nitrogenase.” Integrated Enzyme Database. European Bioinformatics Institute. 22 May 2010. <http://www.ebi.ac.uk/intenz/spotlight.jsp?ec=1.18.6.1>.
7. Noodleman, L., Case, D. A., Han, W.G., Lovell, T., Liu, T., Thompson, M. J., Torres, R. A. “Quantum Bioinorganic Chemistry and Photochemistry.” The Scripps Research Institute. The Scripps Research Institute, 2004. 20 May 2010. <http://www.scripps.edu/news/sr/sr200...noodleman.html>.
8. Ryle, Matthew J., Lanzilotta, William N. Seefeldt, Lance C., Scarrow, Robert C., Jensen, Gerard M. "Circular Dichroism and X-ray Spectroscopies of Azotobacter vinelandii Nitrogenase Iron Protein." The Journal of Biological Chemistry. The American Society of Biochemistry and Molecular Biology, 1996. 23 May 2010. <http://www.jbc.org/content/271/3/1551>.
9. Simmons, Kent. “The Diversity of Life.” Evolution, Ecology, and Biodiversity. 2006. 24 May 2010. <http://kentsimmons.uwinnipeg.ca/16cm...6/16monera.htm>.
10. Tatum, Malcolm. “What is Nitrogenase?” Wisegeek. 22 May 2010. <http://www.wisegeek.com/what-is-nitrogenase.htm>.

