Misc Antibiotics
- Page ID
- 419
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Antibiotics are specific chemical substances derived from or produced by living organisms that are capable of inhibiting the life processes of other organisms. The first antibiotics were isolated from microorganisms but some are now obtained from higher plants and animals. Over 3,000 antibiotics have been identified but only a few dozen are used in medicine. Antibiotics are the most widely prescribed class of drugs comprising 12% of the prescriptions in the United States.
Macrolides
Macrolides are products of actinomycetes (soil bacteria) or semi-synthetic derivatives of them. Erythromycin is an orally effective antibiotic discovered in 1952 in the metabolic products of a strain of Streptocyces erythreus, originally obtained from a soil sample.
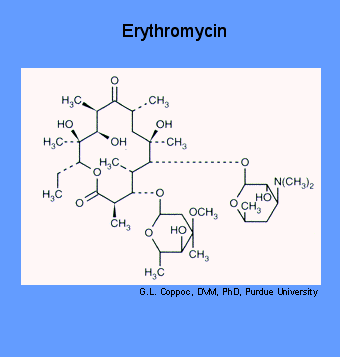
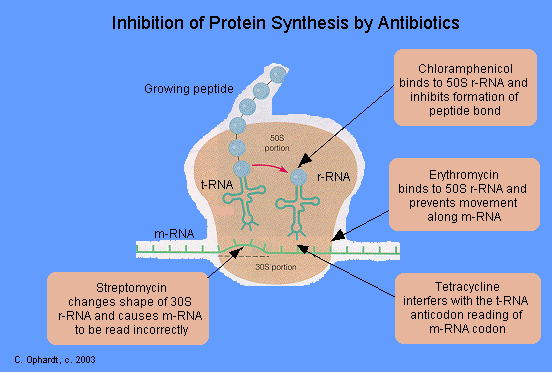
Erythromycin and other macrolide antibiotics inhibit protein synthesis by binding to the 23S rRNA molecule (in the 50S subunit) of the bacterial ribosome blocking the exit of the growing peptide chain. of sensitive microorganisms. (Humans do not have 50 S ribosomal subunits, but have ribosomes composed of 40 S and 60 S subunits). Certain resistant microorganisms with mutational changes in components of this subunit of the ribosome fail to bind the drug. The association between erythromycin and the ribosome is reversible and takes place only when the 50 S subunit is free from tRNA molecules bearing nascent peptide chains. Gram-positive bacteria accumulate about 100 times more erythromycin than do gram-negative microorganisms. The non ionized from of the drug is considerably more permeable to cells, and this probably explains the increased antimicrobial activity that is observed in alkaline pH.
Tetracyclines
Tetracyclines have the broadest spectrum of antimicrobial activity. These may include: Aureomycin, Terramycin, and Panmycin. Four fused 6-membered rings, as shown in the figure below, form the basic structure from which the various tetracyclines are made. The various derivatives are different at one or more of four sites on the rigid, planar ring structure. The classical tetracyclines were derived from Streptomyces spp., but the newer derivatives are semisynthetic as is generally true for newer members of other drug groups.
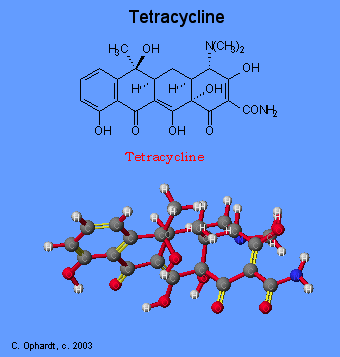
Tetracyclines inhibit bacterial protein synthesis by blocking the attachment of the transfer RNA-amino acid to the ribosome. More precisely they are inhibitors of the codon-anticodon interaction. Tetracyclines can also inhibit protein synthesis in the host, but are less likely to reach the concentration required because eukaryotic cells do not have a tetracycline uptake mechanism.
Streptomycin
Streptomycin is effective against gram-negative bacteria, although it is also used in the treatment of tuberculosis. Streptomycin binds to the 30S ribosome and changes its shape so that it and inhibits protein synthesis by causing a misreading of messenger RNA information.
Chloramphenicol
Chloromycetin is also a broad spectrum antibiotic that possesses activity similar to the tetracylines. At present, it is the only antibiotic prepared synthetically. It is reserved for treatment of serious infections because it is potentially highly toxic to bone marrow cells. It inhibits protein synthesis by attaching to the ribosome and interferes with the formation of peptide bonds between amino acids. It behaves as an antimetabolite for the essential amino acid phenylalanine at ribosomal binding sites.
| Antibiotic | Producer organism | Activity | Site or mode of action |
|---|---|---|---|
| Penicillin | Penicillium chrysogenum | Gram-positive bacteria | Wall synthesis |
| Cephalosporin | Cephalosporium acremonium | Broad spectrum | Wall synthesis |
| Griseofulvin | Penicillium griseofulvum | Dermatophytic fungi | Microtubules |
| Bacitracin | Bacillus subtilis | Gram-positive bacteria | Wall synthesis |
| Polymyxin B | Bacillus polymyxa | Gram-negative bacteria | Cell membrane |
| Amphotericin B | Streptomyces nodosus | Fungi | Cell membrane |
| Erythromycin | Streptomyces erythreus | Gram-positive bacteria | Protein synthesis |
| Neomycin | Streptomyces fradiae | Broad spectrum | Protein synthesis |
| Streptomycin | Streptomyces griseus | Gram-negative bacteria | Protein synthesis |
| Tetracycline | Streptomyces rimosus | Broad spectrum | Protein synthesis |
| Vancomycin | Streptomyces orientalis | Gram-positive bacteria | Protein synthesis |
| Gentamicin | Micromonospora purpurea | Broad spectrum | Protein synthesis |
| Rifamycin | Streptomyces mediterranei | Tuberculosis | Protein synthesis |
Contributors
Charles Ophardt (Professor Emeritus, Elmhurst College); Virtual Chembook

