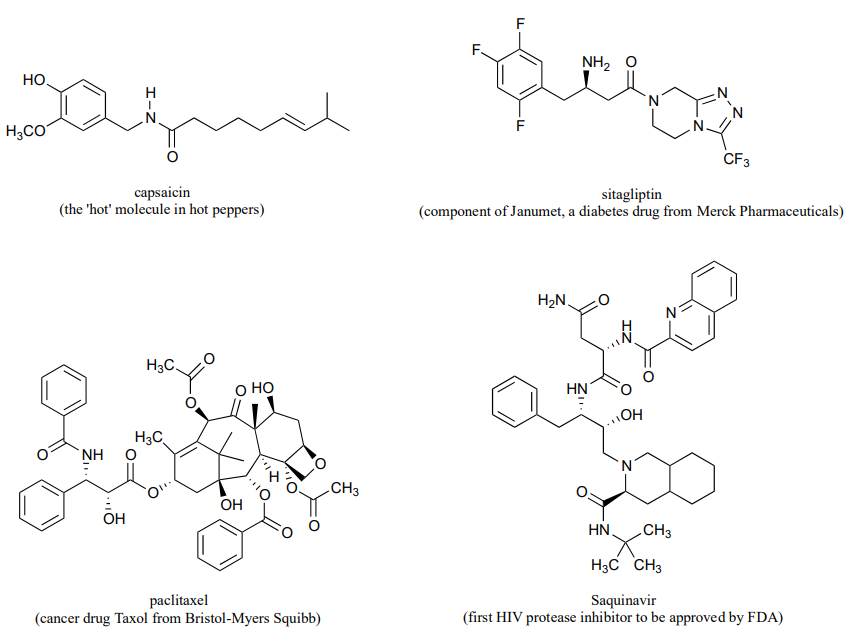
P11.1: Here is some practice in recognizing carboxylic acid derivative functional groups in large, complex biological molecules. There are seven amide and four ester groups in the molecules below - see if you can find them all.
P11.2: (
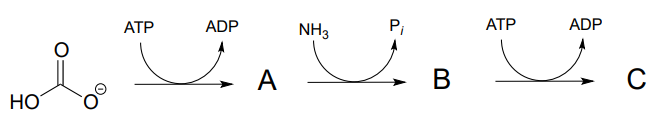
- a) Predict the structures of intermediate compounds A, B, and C in the reaction below (EC 6.3.4.16). Compound C contains an activated carboxylate functionality. Use abbreviations as appropriate.
- Draw a reasonable mechanism for the A to B step
P11.3: Imagine that acetylcholine is combined with acetylcholinesterase ( section 11.6) in a buffer made from \(^{18}O\)-labeled water. Where would you expect to find the \(^{18}O\) label in the products?
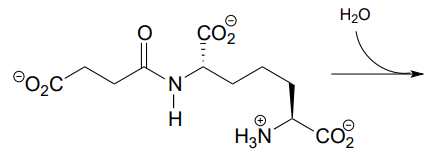
P11.4: Predict the products of this hydrolysis reaction (EC 3.5.1.18):
P11.5: The breakdown of fat in our bodies begins with the action of lipase enzymes, which catalyze the cleavage of fatty acids from the glycerol backbone of triacylglycerol (see section 1.3 for a reminder of the structure of triacylglycerol). A serine residue in the lipase active site plays a key nucleophilic role in the reaction. Draw the single mechanistic step in which the covalent link between a fatty acid and the glyceryl backbone is broken, using curved arrow notation and appropriate abbreviation.
P11.6: Before long-chain fatty acids are transported across the inner mitochondrial membrane, they are temporarily transferred from Coenzyme A to a transport molecule called carnitine, to which they are linked by an ester group (EC 2.3.1.21).
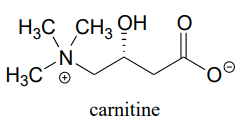
Draw the structure of fatty acyl carnitine (use R to denote the hydrocarbon chain of the fatty acid)
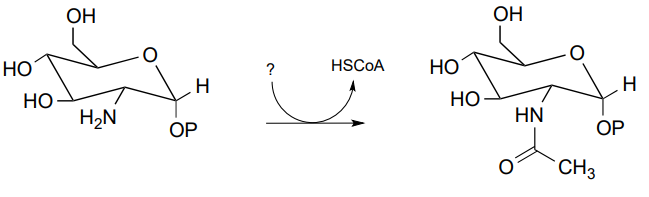
P11.7: Below is a reaction from carbohydrate metabolism (EC 2.3.1.157). Identify the compound designated with a question mark.
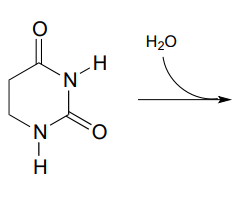
P11.8: Propose the most likely enzymatic hydrolysis product of the substrate below (hint - think about electrophilicity when considering regiochemical outcomes!) (EC 3.5.2.2) J. Biol. Chem. 2006, 281, 13762 scheme 2)
P11.9: The coenzyme tetrahydrofolate (\(THF\)) participates in single-carbon transfer reactions. One derivative of \(THF\), called 10-formyl-\(THF\) (abbreviated structure shown below), transfers a formyl group early in purine ribonucleotide biosynthesis to glycinamide ribonucleotide.
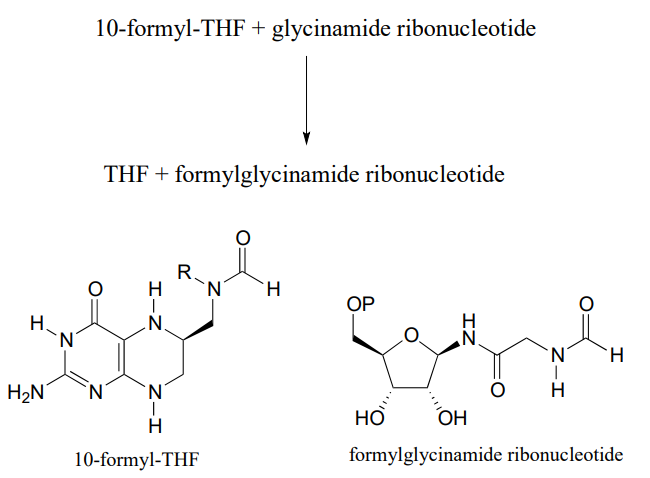
Draw a nucleophilic attack step for this reaction (assume that acyl transfer between the two substrates is direct, without any covalent enzyme-substrate intermediates being formed).
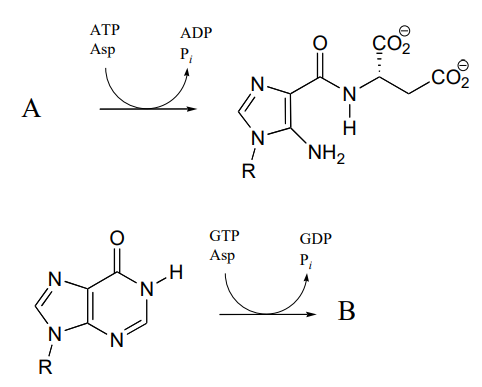
P11.10: One of the key steps in the biosynthesis of purine nucleotides (guanosine and adenosine) in archaea is shown below.
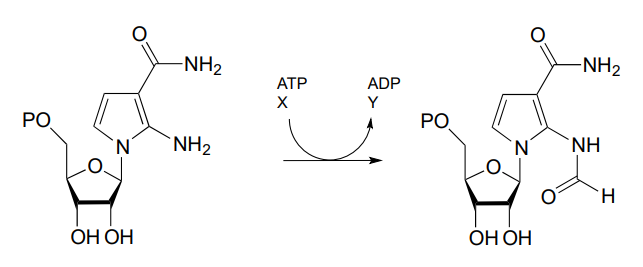
Identify the missing compounds X and Y in the figure above, and draw the structure of an acyl phosphate intermediate.
P11.11: The reactions below are part of nucleotide biosynthesis. Predict the structures of compounds A and B. Compound A contains a carboxylate group, and the reaction that forms compound B is of the type discussed in section 11.8, in which an amine group substitutes at an activated amide to form an amidine/amidinium group.
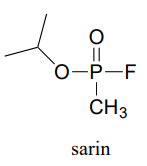
P11.12: Recall from section 11.6 that acetylcholinesterase catalyzes the hydrolysis of the ester group in acetylcholine, going through an intermediate in which the acetyl group on the substrate is transferred to a serine on the enzyme by a transesterification reaction. The nerve gas sarin acts by blocking this initial transesterification step: the drug enters the active site and attaches to the active site serine. Given the structure of sarin below, propose a mechanism for how this happens.
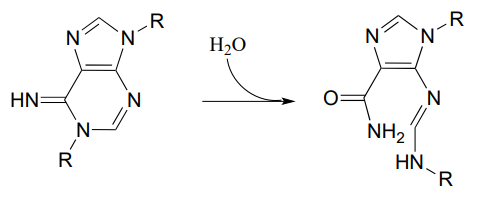
P11.13: Propose a mechanism for the following reaction from histidine biosynthesis (EC 3.5.4.19).
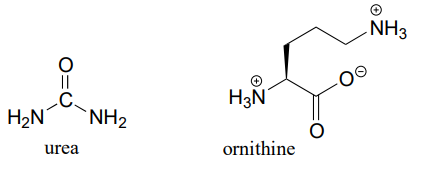
P11.14 (Challenging!) In the final step of the urea cycle (a phase of amino acid degradation pathways), the amino acid arginine is hydrolyzed to urea and ornithine (EC 3.5.3.1). Propose a reasonable mechanism.
P11.15: In the biosynthetic pathway for the DNA/RNA bases uridine and cytidine, a reaction occurs in which carbamoyl phosphate condenses with aspartate, and the resulting intermediate cyclizes to form dihydroorotate. Propose a mechanism for this transformation. Hint: in a very unusual step, a carboxylate group is at one point in the process directly subjected to an acyl transfer reaction, without prior activation by phosphorylation. The enzyme accomplishes this with the help of two bo.und zinc ions, which serve to stabilize the negative charge on a hydroxide leaving group. (EC 3.5.2.3) (Biochemistry 2001, 40, 6989, Scheme 2)
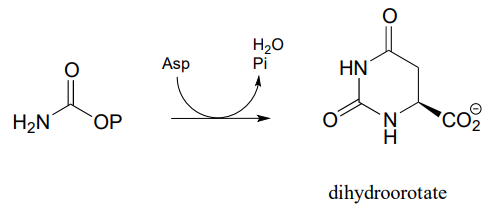
P11.16: Propose a reasonable mechanism for the reaction below (from lysine biosynthesis), and fill in the missing species indicated by question marks.
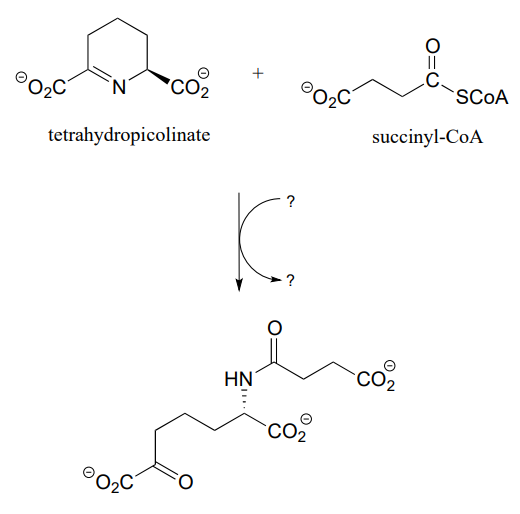
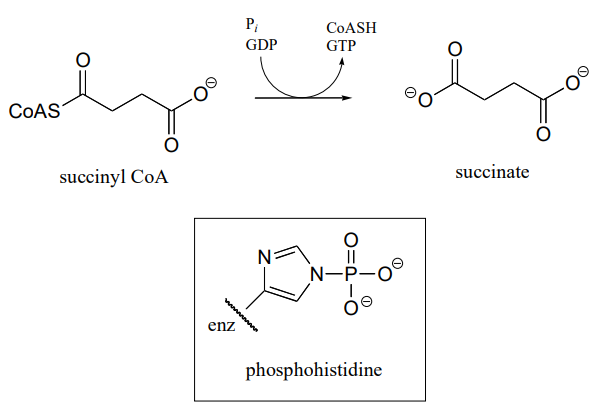
P11.17: In a step in the citric acid cycle, hydrolysis of succinyl-\(CoA\) is coupled to phosphorylation of GDP. The mechanism involves the transient phosphorylation of an active site histidine. Suggest a (multi-step) mechanism for this process (EC 6.2.1.4).
P11.18: A \(^{14}C\)-labeled diazoketone compound (structure below) was used to irreversibly inactivate an enzyme called glutaminase A. Inactivation was shown to occur with \(^{14}C\) labeling of an active site cysteine.
a)Propose a mechanism of inactivation and cysteine labeling.
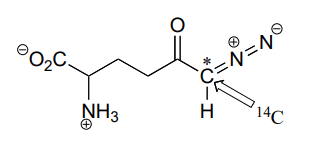
b) To a lesser extent, inactivation of the enzyme and labeling of the cysteine was found to occur with release of a radioactive compound from the active site. Propose a mechanism for the mode of inactivation.
P11.19: Dehelogenase enzymes catalyze the cleavage of carbon-halogen bonds, and are of interest to scientists looking for new ways to detoxify organohalogen pollutants that make their way into the environment. One such dehalogenase catalyzes the following reaction:
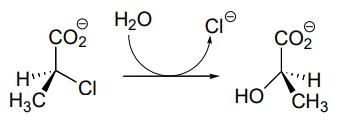
An active site aspartate is thought to carry out the initial nucleophilic attack that expels the chloride.
- Draw a likely mechanism for the complete reaction shown above. Look carefully at the stereochemical progress!
- When the active site aspartate was mutated to asparagine, the enzyme still maintained activity. Mass spectrometry analysis indicated that, at one point in the catalytic cycle of the mutant enzyme, the asparagine exists as a cyanoalanine. Draw a likely mechanism for the reaction as catalyzed by the mutant enzyme, including formation of the transient cyanoalanine residue and subsequent regeneration of the asparagine.



















