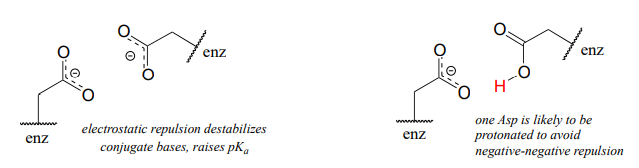Virtually all biochemical reactions take place inside the active site pocket of an enzyme, rather than free in aqueous solution. The microenvironment inside an enzyme's active site can often be very different from the environment outside in the aqueous solvent. Consider, for example, the side chain carboxylate on an aspartate residue in an enzyme. The literature \(pK_a\) of this carboxylic acid group is listed as 3.9, but this estimate assumes that the group is positioned on the surface of the protein, exposed to water. In the physiological buffer of \(pH ~ 7\), a carboxylic acid group with \(pK_a = 3.9\) will be fully deprotonated and negatively charged. If, however, an aspartate side chain happens to be buried deep inside the interior of the protein’s active site, and is surrounded primarily by nonpolar side chains such as alanine, phenylalanine, tryptophan, etc., the situation is very different.
Cut off from the environment of the bulk solvent, the carboxylate group (red in the above figure) is now in a very nonpolar microenvironment, a situation in which the protonated, uncharged state is stabilized relative to the deprotonated, negatively charged state (this is simply another application of the 'like dissolves like' principle you learned in General Chemistry - a charged group is highly destabilized by a nonpolar environment). The overall effect is that the \(pK_a\) for this aspartate residue is actually higher than 3.9 – it is less acidic, and more likely to be in its protonated form inside the protein.
A similar effect would be observed in a situation where the side chain carboxylate groups of two aspartate residues are located in close proximity to one another in an enzyme active site. Two negatively charged groups close to each other represents a very high energy, repulsive situation, and this can be relieved if one of the two side chains is protonated.
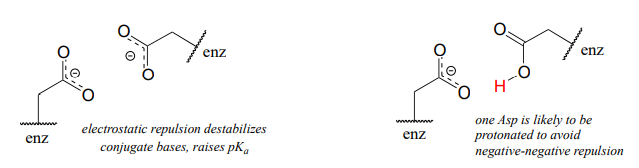
In this microenvironment, the proximity of one amino acid group directly effects the pKa of its neighbor.
Now consider a situation where a metal ion such as magnesium (\(Mg^{+2}\)) or zinc (\(Zn^{+2}\)) is bound in the interior of the enzyme, in close contact with an aspartate side chain. With a cation to interact with, the anionic, deprotonated state of the amino acid is stabilized, so the \(pK_a\) of this Asp residue is likely to be substantially lower than 3.9.
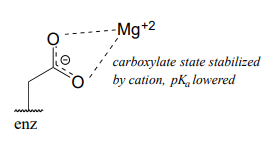
The metal ion in this situation is considered to be acting as a Lewis acid, accepting electron density from the carboxylate group.
The \(pK_a\)-lowering effect of a metal cation can be dramatic – it has been estimated that a water molecule coordinated to a \(Cu^{+2}\) or \(Zn^{+2}\) ion can have a \(pK_a\) as low as 7 (compare this to the ‘normal’ water \(pK_a\) of 15.7!)
A lysine residue located deep in the interior of a protein is surrounded by nonpolar residues. In what direction will this alter the 'normal' \(pK_a\) of the lysine side chain, and why?
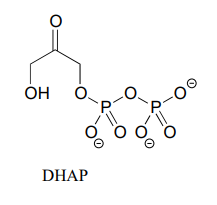
In many biochemical reactions which involve the formation of an enolate intermediate, the carbonyl oxygen of the substrate is coordinated to a divalent metal ion (usually zinc or magnesium) in the active site. Explain, with structural drawings, how this ion-dipole interaction effects the acidity of the \(\alpha \)-protons of dihydroxyacetone phosphate (DHAP), an intermediate compound in the glycolysis pathway.
Video tutorials: organic acid-base chemistry