4.1: Myoglobin, Hemoglobin, and their Ligands
- Page ID
- 165191
Almost all biochemistry textbooks start their description of the biological functions of proteins using the myoglobin and hemoglobin as exemplars. These are very rational approaches since they have become model systems to describe the binding of simple ligands, like dioxygen (O2), CO2, and H+, and how the structure of the protein determines and is influenced by binding of ligands.
Yet in a way these "ligands" are dissimilar to perhaps the majority of other proteins which bind small ligands such as substrates (for enzymes), inhibitors and activators or large "ligands" such as other proteins, nucleic acids, carbohydrates and lipids. These type of ligands are reversibly bound through classical intermolecular forces (IMFs), such as hydrogen bonds, London dispersion forces, dipole-dipole interactions, and ion-ion interactions. In addition to these are the less commonly discussed pi-pi (aromatic) interactions and cation-pi (aromatic) interactions.
![]() YouTube: Less Common Discussed IMFs - pi-pi, cation-pi interactions, and halogen bnding
YouTube: Less Common Discussed IMFs - pi-pi, cation-pi interactions, and halogen bnding
The classic ligands that reversibly bind to hemoglobin, dioxygen, carbon dioxide, and protons, are bound covalently. Dioxygen binds to a heme Fe2+, protons obviously bind to proton acceptors (like His), while CO2 binds covalently as if forms a carbamate with the N terminus of one of the hemoglobin chains.
This discrepancy in ligand binding mode can be explained easily for dioxygen as it forms a coordinate or dative covalent bond with the transition metal ion Fe+2. In ordinary covalent bonds, each bonded atom contributes to and shares the two electron in the bond. In coordinate or dative covalent bonds, the ligand, a Lewis base, contributes both electrons in the bond. Both electrons are still considered to be "owned" by the ligand and not by the transition metal ion, a Lewis acid. Hence the ligand can readily dissociate from the metal ion, much as a ligand bound through classical IMFs can. This analogy can be extended to protons which are also Lewis acids (with no contributing electrons) as they react with Lewis bases (lone pair donors) on atoms such as N on a His side chain.
Mb (a monomer containing 8 α−helices, A-H) and Hb (a heterotetramer with two α -and two β−subunits, each which also contains 8 α−helices) are both oxygen binding proteins. Dioxygen is transported from lungs, gills, or skin of an animal to capillaries, where it can be delivered to respiring tissue. It has a low solubility in blood (0.1 mM). Whole blood, which contains 150 g Hb/L, can carry up to 10 mM dioxygen. Invertebrate can have alternative proteins for oxygen binding, including hemocyanin, which contains Cu and hemerythrin, a non-heme protein. On binding dioxygen, solutions of Hb change color to bright red. Solutions of hemocyanin and hemerythrin change to blue and burgundy colored, respectively, on binding dioxygen. Some Antarctic fish don't require Hb since dioxygen is more soluble at low temperature. Mb is found in the muscle, and serves as a storage protein for oxygen transported by Hb. Some information about the proteins is given below:
Hemoglobin:
- 1st. protein whose molecular weight was determined
- 1st protein to be assigned a specific function - dioxygen transport
- has a prosthetic group (non-amino acid) heme group (protoporphyrin IX with a ferrous ion) covalently attached to the protein. The heme group binds dioxygen.
- 1st protein in which a point mutation (single base pair charge) causes a single amino acid change in the protein, marking the start of molecular medicine
- 1st protein with high resolution x-ray structure
- theory for dioxygen binding explain control of enzyme activity
- the binding of dioxygen is regulated by binding of H+, CO2, and bisphosphoglcyerate which bind to sites (allosteric) distant from oxygen binding site.
- crystals of deoxy-Hb shatter on binding dioxygen, indicating significant conformational changes on binding.
Heme group:
Figure: Heme
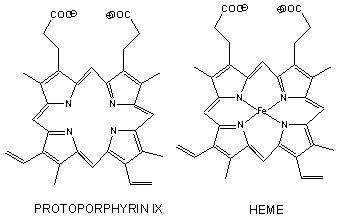
- The heme group contains protoporphyrin IX, with four tetrapyrrole rings linked by methene bridges. Attached to the tetrapyrrole structure are four methyl, two vinyl, and two proprionate groups. These can be arranged in 15 ways, only one (IX) occurs in biological systems. Ferrous ion bonded to the protoporphyrin IX constitutes the heme group.
- The heme fits into a hydophobic crevice in the proteins with the proprionate groups exposed to solvent.
- The Fe2+ ion is coordinated to 4 N's on the 4 pyrrole rings, The 5th ligand is a supplied by proximal His (the 8th amino acid on helix F) of the protein. In the absence of dioxygen, the 6 ligand is missing. and the geometry of the complex is square pyramidal with the Fe above the plane of the heme ring. A distal His (E7) is on the opposite side of the heme ring, but too far to coordinate with the Fe. When dioxgen binds, it occupies the 6th coordination site and pulls the Fe into the plane of the ring, leading to octahedral geometry. CO, NO, and H2S also bind to the 6th site, but with higher affinity than dioxygen, which can lead to CO poisoning. The distal His keeps these ligands (including dioxygen) bound in a bent, non-optimal geometry. This minimizes the chances of CO poisoning.
- When oxygen binds to heme Fe2+, the geometry of the complex becomes octahedral. The Fe2+ has 6 electrons in d orbitals. (The electronic configuration of Fe is 3d64s2 while the Fe2+ ion is 3d6). From crystal field theory (review a general chemistry text), the ligand electrons pairs (treated as anions) interact with the d orbital Fe2+ electrons. They interact most strongly (in a repulsive fashion) with the dz2 and dx2-y2 Fe2+ electrons, which are oriented at the axial ends and equatorial corners, respectively, of the octahedral complex. This repulsion raises the energy of these two orbitals. The other three d orbitals feel less repulsion by the ligand electrons and are lower in energy. The difference in energy between the three lower energy levels of d electrons (in t orbitals) and the higher energy levels of electrons (in g orbitals) allows photons matching that energy difference to be absorbed as electrons are promoted to the g orbitals. Ligands that interact strongly with the dz2 and dx2-y2 electrons raise their energy even more, increasing the energy of the photon required to promote an electron from the t to g orbitals. Dioxygen is one of those ligands, leading to the absorbance of high energy photons, causing the solution to appear bright red (since blue light is absorbed). Because of this strong ligand, the significant energy gap between the t and g orbitals causes the electrons to reside mostly in the lower energy t orbitals where they remained paired, giving the complex a low spin state in which all 6 d electrons are paired. Hence oxyhemoglobin is diagmagnetic. The heme complex can also exist in a high spin state in which the electrons are easily excited to the g orbitals which are closer in energy to the t orbitals when the ligand is weakly interacting with the Fe2+ electrons. In these cases, the heme is paramagnetic since unpaired electrons are possible. (We will discuss paramagnetism latter in the course during our exploration of oxygen chemistry.
Figure: Heme-O2 Octahedral Complex
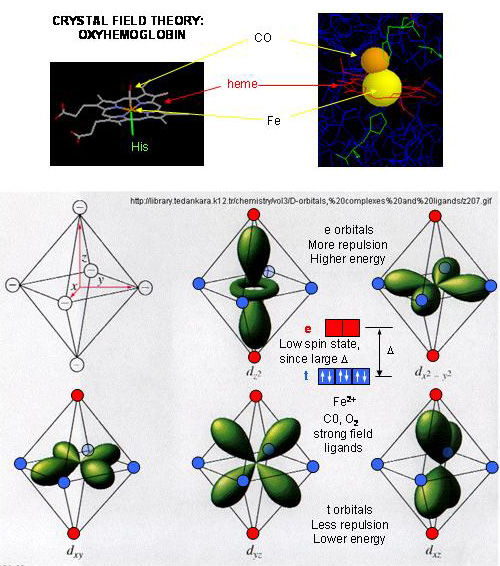
- When dioxyen binds, the oxidation state of the Fe2+ ion does not change, even through dioxygen is a great oxidizing agent. Hence the Fe2+ ion is a reversible carrier of dioxygen not of electrons. Free heme in solution is oxidized by dioxygen, forming a complex with water which occupies the 6th position, with the iron in the Fe3+ state. An intermediate in this process is the formation of a dimer of 2 hemes linked by 1 dioxygen. This can't occur readily when the heme is in Hb or Mb. Other heme proteins (like Cytochrome C) are designed to be carriers of electrons.
- A small amount of the Fe2+ ion gets oxidized to Fe3+ ion. Mb and Hb in this state are called metHb and metMb. A enzyme is required to reduced the iron back to the Fe2+ state.
![]() Jmol: Updated deoxy-heme and carbon monoxide-heme Jmol14 (Java) | JSMol (HTML5)
Jmol: Updated deoxy-heme and carbon monoxide-heme Jmol14 (Java) | JSMol (HTML5)
Myoglobin
- Mb is extremely compact, and consists of 75% alpha helical structure.
- 8 α−helices are present, labeled A-H.
- 4 alpha helices are terminated by Pro
- The interior amino acids are almost entirely nonpolar. The only polar amino acids found completely buried are the two His (proximal and distal) found at the active site of dioxygen binding.
![]() Jmol: Updated Met-Myoglobin Jmol14 (Java) | JSMol (HTML5)
Jmol: Updated Met-Myoglobin Jmol14 (Java) | JSMol (HTML5)
Difference between Hb and Mb
- Hb is a tetramer of two αand two β subunits held together by IMF's (an example of quarternary protein structure), and 4 bound hemes, each of which can bind a dioxygen. In a fetus, two other subunits make up Hb (two zeta - ζand two epsilion - ε subunits -analogous to the two αand two β subunits, respectively). This changes in fetuses to two α and two γ subunits. Fetal Hb has higher affinity for dioxygen than adult Hb. Mb is a single polypeptide chains which has higher affinity for dioxygen than Hb.
The α and β chains are similar to Mb, which is unexpected since only 24 of 141 residues in the αand β chains of Hb are identical to amino acids in Mb. This suggests that different sequences can fold to similar structures. The globin fold of Mb and each chain of Hb is common to vertebrates and must be nature's design for dioxygen carriers. A comparison of the sequence of Hb from 60 species show much variability of amino acids, with only 9 identical amino founds. These must be important for structure/function. All internal changes are conservative (e.g. changing a nonpolar for a nonpolar amino acid). Not even Pro's are conserved, suggesting there are different ways to break helices. The two active site His are conserved, as is Gly B6 (required for a reverse turn). http://www.umass.edu/molvis/tutorials/hemoglobin/
![]()
![]() Jmol: Hemoglobin
Jmol: Hemoglobin


