13.2: HIV-1 protease (PR)
- Page ID
- 169622
Proteases are enzymes that have the ability to cut proteins into peptides. In order for maturation of HIV to occur, a HIV enzyme termed a protease has to cleave a long HIV-encoded gag-pol polyprotein to produce reverse transcriptase and integrase (coded by the HIV pol gene) and gag polyprotein (coded by the HIV gag gene). The HIV protease then cleaves the gag polyprotein into capsid protein p17, matrix protein p24, and nucleocapsid protein p7, as well as proteins p6, p2, and p1 whose functions are not yet fully understood. Proteases also cleave the env-polyprotein (coded by the HIV env gene) into the envelope glycoproteins gp120 and gp41. This allows the completion of the assembly step in the viral life cycle where the proteins and the viral RNA come together to form virion particles ready to exit the cell.
HIV-1 protease (PR) Inhibitor
Protease inhibitors are short peptide-like molecules that are competitive inhibitors of the enzyme. Instead of -NH-CO- peptide link, they contain -(CH2-CH(OH)-). When such a peptide gets into the enzyme active site, the protease is unable to cut the linkage and gets inactivated. This leads to a lack of cleavage of the polypeptide chains of two crucial viral proteins, Gag and Pol, which are essential structural and enzymatic proteins of HIV. Their absence blocks the formation of mature virion particles.
Mutations in the enzyme active site and other sites, which cause conformational changes, can cause resistance. Quite often one mutation can lead to resistance to many different drugs simultaneously since they all share the same mode of action. This is called cross-resistance. It is one of the major drawbacks of protease inhibitors therapy.
Mechanism of HIV-1 protease
The mechanism of HIV-1 protease is still yet to be fully understood. The main way the mechanism are studied is through the use of mimicry substrates and simulations. HIV-1 protease has been studied intensely using various inhibitors, observing partial steps of the process. Since the main target of these inhibitors is to bind to the Asp-25 of the catalytic triad, each inhibitor would vary in its mechanism to accomplish this.
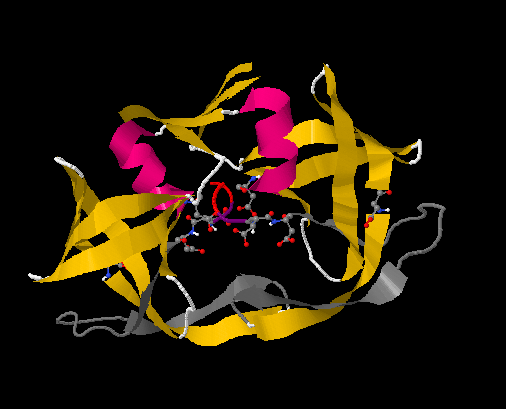
The first part of the mechanism begins with the substrate binding onto the protease. Figure 13.2.1 accents the key amino acids in HIV-1 protease that assists in substrate binding. It is predicted that a substrate first binds via a hydrogen bond to aspartic acid 30 on one chain. Once this initial bond is made, the binding is then further stabilized by bondage to the glycine rich region in the flap of the same monomer. A salt bridge is then formed from the substrate to glutamic acid 35 of the other monomer. This completes binding of the substrate to the protease. At this point, waters molecules that are found at the tips of the flaps at isoleucine 50 on each monomer dissociates from the protease. The release of the water molecule results in a structural conformation change of the protease, changing it from semi-open to closed, tightening the space between the protease and substrate.
Once in the tightened state, aspartic acid 25 and 25’ hydrogen binds to their adjacent glycine, and then becomes supported by the following threonine. Originally, there is a water molecule bound between the aspartic acids. One of the aspartic acid exists in a deprotonated state and the other one is protonated. The water molecule stabilizes the aspartates in this form.When the substrate binds to the protease, it causes conformational changes that brings the substrate to the position of the water molecule, and the water molecule acts as a nucleophile to the substrate. The oxygen of the water attacks the carbonyl group of the substrate peptide bond that is by the active site as the nitrogen picks up the hydrogen of the protonated aspartic acid. What results is an hydroxl group is added to the carbonyl group as an amine is formed on other side of the peptide bond, leaving a hydrogen atom behind to stablize the two aspartates. This is proposed to occur in a concerted fashion.
Contributors
Curation and Revision. Provided by: Boundless.com. License: CC BY-SA: Attribution-ShareAlike

