5.5: Mass Spectrometry
- Page ID
- 320018
A final technique in structure elucidation is mass spectrometry, which allows us to measure the mass of an ion via a mass-to-charge (m/z) ratio.
The basic schematic of a mass spectrometer is shown in Figure. 5.5.1:
There are four main components:
- Ionization: After injection of a sample and vaporization via the sample inlet, the sample is ionized by one of several mechanisms.
- Acceleration: The vaporized beam of ions is accelerated so that they have the same energy
- Deflection: In the mass analyzer (an electromagnet), the ions are sorted according to their size (m/z).
- Detection: Ions finally strike a plate and generate a current
The type of mass spectrometer is normally determined by the mechanism of ionization:
Electron Impact (EI) - high energy electrons (6700 kJ/mol) are bombarded at a neutral molecule. The resulting high energy radical cation can break apart into smaller fragments (this is called fragmentation - the molecular ion (M+) breaks down into small daughter peaks). Because electron impact is a high energy technique, one often does not see the molecular ion M+, only daughter peaks due to rapid fragmentation.
Chemical Ionization (CI) - a cloud of high energy methane (CH4) gas and a source of protons (H+ ions) transfer a proton to a small molecule to generate a molecular ion [M+H]+ (observed in the mass spectrum at a m/z ratio of M+1).
Electrospray ionization (ESI) - the most common technique droplets of a small molecule in solvent are created via an aerosol from the sample inlet. Normally, the solvent contains a proton source which protonates basic sites on the small molecule. As the solvent shell evaporates, a charged small molecule results.
Matrix-Assisted Laser Desorption Ionization (MALDI) - a sample is mixed with a chemical matrix and deposited on a plate to dry and crystallize. In a vacuum chamber, the plate is pulsed with a laser that ionizes and desorbs ions, which are now in the gaseous state, usually [M+H]+ or [M+Na]+.
Isotopic patterns
The natural abundance of certain isotopes can be useful in structure elucidation. For example, we can tell if a molecular ion has a chlorine or bromine atom in it just by loking at the isotopic pattern of the molecule ion or daughter peaks. To do this, we need a high resolution mass spectrum (HRMS). HRMS is necessary for accurate mass determination, and we use the following exact masses:
12C = 12.000000 amu
1H = 1.007825 amu
16O = 15.99491
14N = 14.00307
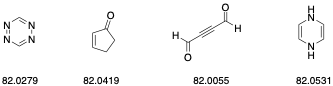
For calculating the mass-to-charge ratio of an ion, just add up all of the masses of the atoms in the ion using these values. Since these values are extremely accurate, only a handful (or maybe only one) of molecules can have that exact mass, down to an accuracy of less than 10 ppm. For example, we can distinguish each of the molecules below, which have a molecular weight of 82 g/mol, via HRMS (using EI).
Getting back to isotopic patterns, we see that for the following molecule, there are two molecular ions because the natural abundance of chlorine is 35Cl (75%) and 37Cl (25%). Thus, is you have a sample of the molecule, 75% of it will have 35Cl atoms and 25% of it will have 37Cl atoms. You will see both bypes of species in a mass spectrum in a 3:1 ratio separated by two mass units.
Other typical patterns in mass spectra are when bromine atoms appear. Here, one observes peaks that are two mass units apart and equal in intensity. This is because 79Br and 81Br are naturally abundant in a 1:1 ratio.
Let's see what would happen if there were multiple halogen atoms, as in CH2Cl2 (MW = 85 g/mol).
Peak 1: m/z 84 = CH235Cl35Cl = (0.75)*(0.75) = 56.25 = 56.25%
Peak 2: m/z 86 = 37.5%
(second chlorine is 37Cl) = CH235Cl37Cl = (0.75)*(0.25) = 18.75 = 18.75%
(first chlorine is 37Cl) = CH237Cl35Cl = (0.75)*(0.25) = 18.75 = 18.75%
Peak 3: m/z 88 = CH237Cl37Cl = (0.25)*(0.25) = 6.25 = 6.25%
Biomedical Spotlight
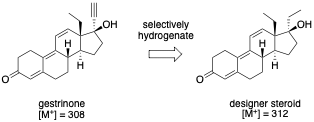
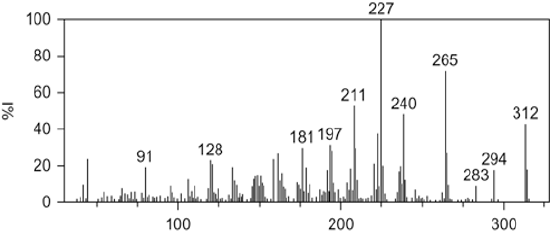
The United States Anti-Doping Agency (USADA) is the nation's premier non-profit organization charged with upholding the ideal of fair play by ensuring that athletic competitions are free from performance-enhancing drugs. In 2003, USADA was sent a syringe containing an unknown substance from an anonymous coach who claimed that there had been rampant use of a designer steroid among track and field athletes. In what then came widely known as the BALCO scandal, this same substance was implicated in Major League Baseball players, including Barry Bonds. A sample from the syringe was tested for all 70 steroids that are banned by USADA, but no matches were found. However, the mass spectrum had remarkable similarity to a known anabolic agent, gestrinone. In fact, the daughter peaks from electron impact ionization mass spectrometry (EI-MS) for a sample of the designer steroid completely matched that of gestrinone. However, the molecular ion for the designer steroid differed by 4 amu (EI-MS shown below). By using chemical intuition, anti-doping scientists were able to conclude that gestrinone had been partially and selectively hydrogenated to produce an ethyl group from the terminal alkyne. This new molecule had performance-enhancing capabilities, but had previously escaped detection. The new molecule, now known as tetrahydrogestrinone (THG), was quickly banned by the World Anti-Doping Agency.