5.4: Simulating DNA's Diffraction Pattern
- Page ID
- 150519
The publication of the DNA double-helix structure by x-ray diffraction in 1953 is one the most significant scientific events of the 20th century (1). Therefore, it is important that science students and their teachers have some understanding of how this great achievement was accomplished. X-ray diffraction is conceptually simple: a source of X-rays illuminates a sample which scatters the x-rays, and a detector records the arrival of the scattered x-rays (diffraction pattern). However, the mathematical analysis required to extract from diffraction pattern the molecular geometry of the sample that caused the diffraction pattern is quite formidable. Therefore, the purpose of this tutorial is to illustrate some of the elements of the mathematical analysis required to solve a structure.
The famous X-ray diffraction pattern obtained by Rosalind Franklin is shown below (2).
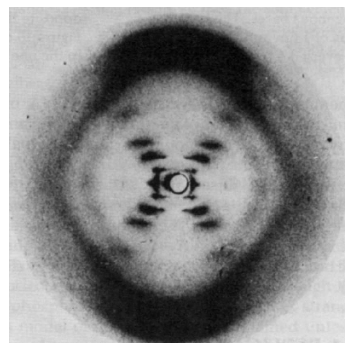
This X-ray picture stimulated Watson and Crick to propose the now famous double-helix sturcture for DNA. It was surely fortuitous that Crick had recently completed an unrelated study of the diffraction patterns of helical molecules (3).
To gain some understanding of how the experimental pattern led to the hypothesis of a double-helical structure we will work in reverse. We will assume the double-helix structure, calculate the diffraction pattern, and compare it with the experimental result. This, therefore, is a deductive exercise as opposed to the brilliant inductive accomplishment of Watson and Crick in determining the DNA structure from Franklin's experimental X-ray pattern.
The experimental pattern will be simulated by modeling DNA solely as a planar double strand of sugar-phosphate backbone groups shown below. Reference 4 provides the justification and the limitations in using two-dimensional models for three-dimensional structures when simulating X-ray diffraction experiments.
The double-strand geometry shown below was created using the following mathematics. Calculations are carried out in atomic units.
\[ \begin{matrix} \text{Sugar-phosphate groups per strand:} & A = 20 & \text{Strand radius:} & R = 1 & \text{Phase difference between strands:} & 0.8 \pi \end{matrix} \nonumber \]
\[ \begin{matrix} \text{First strand:} & m = 1 .. A & \Theta_m = \frac{4 \pi m}{A} & y_m = m & x_m = R \cos \left( \Theta_m \right) \\ \text{Second strand:} & m = 21 .. 40 & \Theta_m = \frac{4 \pi (m - A)}{A} & y_m = (m-A) & x_m = R \cos \left( \Theta_m + 0.8 \pi \right) \\ m = 1 .. 20 & n = 21 .. 40 \end{matrix} \nonumber \]
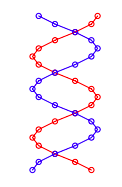
According to quantum mechanical principles, the photons illuminating this geometrical arrangement interact with all its members simultaneously thus being cast into the spatial superposition, Ψ, given below.
\[ | \Psi \rangle = \frac{1}{ \sqrt{N}} \sum_{i = 1}^N |x_i,~y_i \rangle \nonumber \]
This spatial wave function is then projected into momentum space by a Fourier transform to yield the theoretical diffraction pattern. What is measured at the detector according to quantum mechanics is the two-dimensional momentum distribution created by the spatial localization that occurs during illumination of the structure. If the sugar-phosphate groups are treated as point scatterers the momentum wave function is given by the following Fourier transform.
\[ \Theta (p_x,~p_y) = \frac{1}{2 \pi} \sum_{m = 1}^{40} exp (-i~p_x~x_m ) exp( -i~p_y~y_m) \nonumber \]
The theoretical diffraction pattern can now be displayed as the absolute magnitude squared of the momentum wave function.
\[ \begin{matrix} \Delta = 8 & N = 200 & j = 0 .. N & px_j = - \Delta + \frac{2 \Delta j}{N} & k = 0 .. N & py_k = - \Delta + \frac{2 \Delta k}{N} \end{matrix} \nonumber \]
\[ \text{Diffraction pattern}_{j,~k} = \left( \left| \Phi \left( px_j,~y_k \right) \right| \right)^2 \nonumber \]
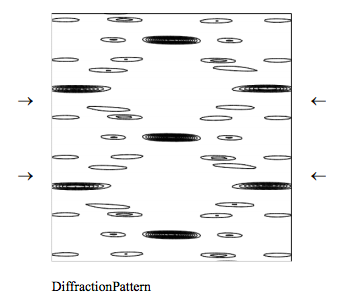
Clearly the naive model diffraction pattern presented here captures several important features of the experimental diffraction pattern. Among those are the characteristic X-shaped cross of the diffraction pattern and the missing fourth horizontal layer (indicated by arrows).
Lucas, Lisensky, and co-workers (4, 5) have simulated the DNA diffraction pattern using the optical transform method. This tutorial might therefore be considered to be a theoretical companion to their more empirical approach to the subject.
References:
- Watson, J. D.; Crick, F. H. C. Nature 1953, 171, 737.
- Franklin, R. E.; Gosling, R. G. Nature 1953, 171, 740.
- Cochran, W.; Crick, F. H. C.; Vand, V. Acta Crystallogr. 1952, 5, 581.
- Lucas, A. A.; Lambin, Ph.; Mairesse, R.; Mathot, M. J. Chem. Educ. 1999, 76, 378.
- Lisensky, G. C.; Lucas, A. A.; Nordell, K. J.; Jackelen, A. L.; Condren, S. M.; Tobe, R. H.; Ellis, A. B. DNA Optical Transform Kit; Institute for Chemical Education: University of Wisconsin, WI, 1999.

