13.2: Decarboxylation
- Page ID
- 106372
Many carbon-carbon bond-forming and bond-breaking processes in biological chemistry involve the gain or loss, by an organic molecule, of a single carbon atom in the form of \(CO_2\). You undoubtedly have seen this chemical equation before in an introductory biology or chemistry class:
\[\ce{CO2 (g) + 6H2O (l) + energy \rightarrow C6H12O6 (aq) + 6O2 (g)}\]
This of course represents the photosynthetic process, by which plants (and some bacteria) harness energy from the sun to build glucose from individual carbon dioxide molecules. The key chemical step in plants in which a carbon dioxide molecule is 'fixed' (linked to a larger organic molecule) is a carboxylation reaction, and is catalyzed by the enzyme ribulose 1,5-bisphosphate carboxylase, commonly known as Rubisco.
The reverse chemical equation is also probably familiar to you:
\[\ce{C6H12O6(aq) + 6O2(g) \rightarrow 6CO2(g) + 6H2O(l) + energy}\]
This equation expresses what happens in the process known as respiration: the oxidative breakdown of glucose to form carbon dioxide, water, and energy (in a non-biological setting, it is also the equation for the uncatalyzed combustion of glucose). In respiration, each of the carbon atoms of glucose is eventually converted to a \(CO_2\) molecule. The process by which a carbon atom - in the form of carbon dioxide - breaks off from a larger organic molecule is called decarboxylation.
We will look now at the biochemical mechanism of decarboxylation reactions. Later in the chapter, we will look at the carboxylation reaction catalyzed by the Rubisco enzyme.
Decarboxylation steps occur at many points throughout central metabolism. Most often, the substrate for a decarboxylation step is a \(\beta\)-carboxy ketone or aldehyde.
Decarboxylation of a b-carboxy ketone or aldehyde:
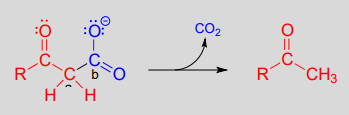
Mechanism:
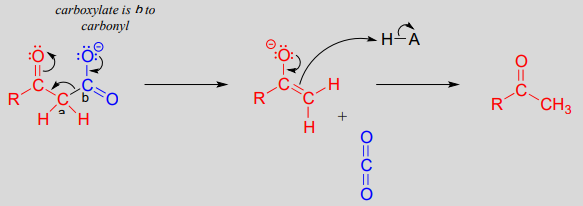
Just as in a retro-aldol reaction, a carbon-carbon bond is broken, and the electrons from the broken bond must be stabilized for the step to take place. Quite often, the electrons are stabilized by the formation of an enolate, as is the case in the general mechanism pictured above.
The electrons from the breaking carbon-carbon bond can also be stabilized by a conjugated imine group and/or by a more extensively conjugated carbonyl.
The key in understanding decarboxylation reactions is to first mentally 'push' the electrons away from the carboxylate group, then ask yourself: where do these electrons go? If the electrons cannot 'land' in a position where they are stabilized, usually by resonance with an oxygen or nitrogen, then a decarboxylation is very unlikely.
The compound below is not likely to undergo decarboxylation:
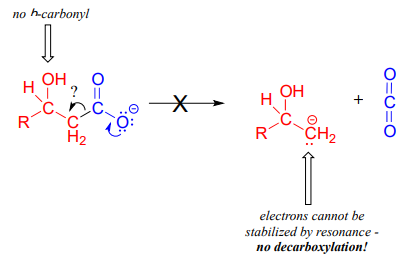
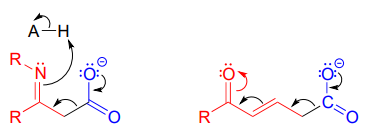
Be especially careful, when drawing decarboxylation mechanisms, to resist the temptation to treat the \(CO_2\) molecule as the leaving group in a mechanistic sense:
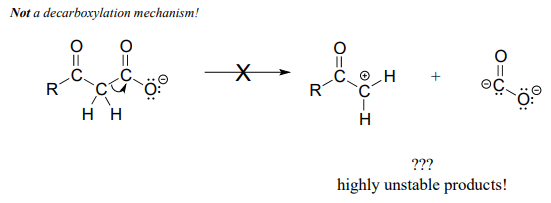
The above is not what a decarboxylation looks like! (Many a point has been deducted from an organic chemistry exam for precisely this mistake!) Remember that in a decarboxylation step, it is the rest of the molecule that is, in fact, the leaving group, 'pushed off' by the electrons on the carboxylate.
Decarboxylation reactions are generally thermodynamically favorable due to the entropic factor: one molecule is converted into two, one of which is a gas - this represents an increase in disorder (entropy). Enzymatic decarboxylation steps in metabolic pathways are also generally irreversible.
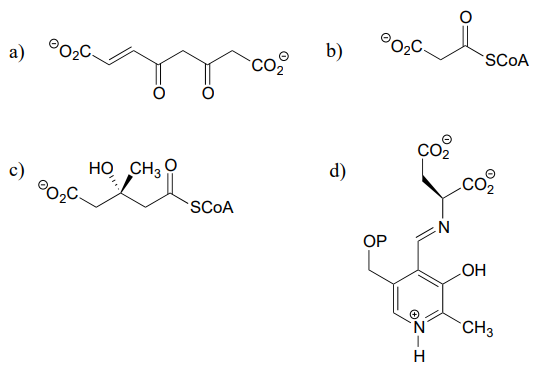
Below are two decarboxylation steps (EC 1.1.1.42; EC 1.1.1.43) in central catabolic metabolism (specifically the citric acid cycle and pentose phosphate pathway catabolism, respectively). Each step representing a point at which a carbon atom derived from the food we eat is released as carbon dioxide:

Draw mechanistic arrows showing the carbon-carbon bond breaking step in each of the reactions shown above.
The reaction catalyzed by acetoacetate decarboxylase (EC 4.1.1.4) relies on an imminium (protonated imine) link that forms temporarily between the substrate and a lysine residue in the active site, in a strategy that is similar to that of the enamine-intermediate aldolase reactions we saw in chapter 12. (Recall from section 7.5 that the \(pK_a\) of an imminium cation is approximately 7, so it is generally accurate to draw either the protonated imminium or the neutral imine in a biological organic mechanism).
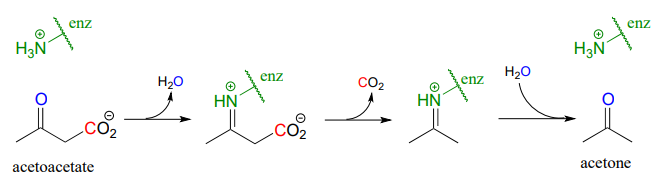
Draw a mechanism for the carbon-carbon bond breaking step in the acetoacetate decarboxylase reaction.
Which of the following compounds could be expected to potentially undergo decarboxylation? Draw the mechanistic arrows for the decarboxylation step of each one you choose, showing how the electrons from the breaking carbon-carbon bond can be stabilized by resonance.