7.6: Complex V
- Page ID
- 190050
Oxidative phosphorylation is a metabolic process in which energy is harnessed for the production of ATP. So far we have looked at the electrons transport chain,which is reponsible for the establishment of a proton gradient across the inner mitochondrial membrane. The actual synthesis of ATP occurs in complex V.
Complex V is very large and contains many different proteins. However, we will simplify the picture here. Schematically, Complex V consists of a proton channel leading from the intermembrane space into the matrix. As protons rush back through the membrane after having been pumped out along the electron transport chain, they cause rotation of a nanoscale millwheel. This millwheel helps bind ADP to phosphate, forming ATP.
- Rotation of a molecular rotor in Complex V helps bring an ADP and a phosphate together to form ATP.
- Rotation of the rotor is powered by protons dropping back through Complex V and into the mitochondiral matrix below.
- The intermembrane space is p-doped (excess positive charge) whereas the mitochondrial matrix is n-doped (excess negative charge), driving the flow of protons toward the mitochondrial matrix.
- For every three protons that drop through the complex, one ATP molecule is formed.
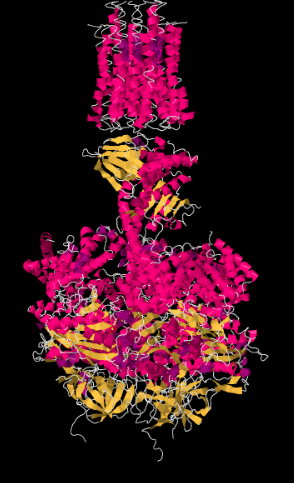
An X-ray crystal structure of Complex V is shown below. At the top of the picture, you can see the membrane-bound portion. Those α-helices run parallel to the fatty acid chains in the lipid layer, so that the protein fits snugly into the membrane. The lower part of the structure is the portion that extends into the matrix.
Exercise \(\PageIndex{1}\)
Recall that in an α-helix, the amino acid residues project outward around the helix. Suggest some residues that are likely to be found along the membrane-bound portion of the complex.
- Answer
-
These amino acids would probably be non-polar: alanine, glycine, methionine, isoleucine, leucine, methionine, phenylalanine, tryptophan, valine.
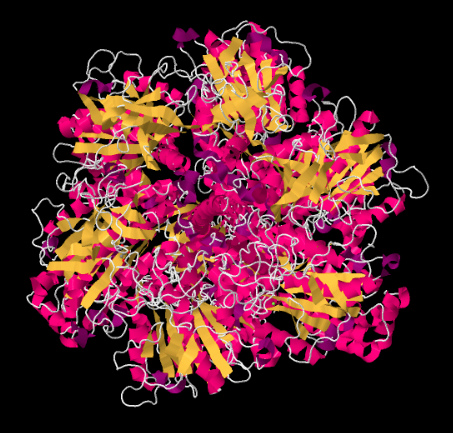
Looking at the structure from underneath -- that is, from the vantage point of the matrix -- we can see the rotational symmetry of Complex V. Complex V contains a molecular rotor that is used in the manufacture of ATP. This rotor is made from a group of proteins that are able to move past another group of proteins as protons pass through the complex.
The mechanism of the rotor works something like an electric motor. The molecular engine contains two parts: a stationary part, or stator, and a rotating part, or rotor. The two parts have complementary charges. The stationary part has a positively charged arginine residue. The rotating part has a negatively charged aspartate residue. The aspartate residue is attracted to the arginine residue, causing the rotor to begin to rotate.
- The stator is the stationary part of a motor.
- The rotor is the revolving part of a motor.
- A temporary attraction draws a site on the rotor toward the stator.
- When that attraction is turned off, momentum carries that site on the rotor past the stator.
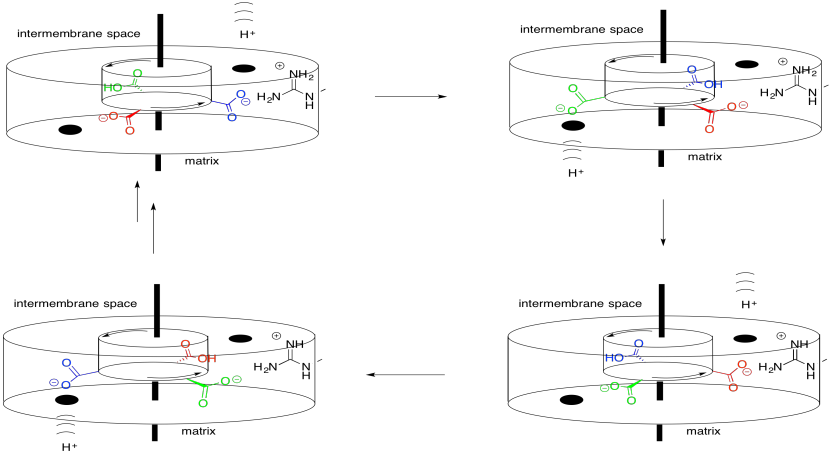
Of course, if that were the entire story, the aspartate (and the rotor) would stop moving when it got as close as possible to the arginine. That would be the end of the story. Instead, a proton arrives as it moves from the intermembrane space toward the matrix. It binds to the aspartate, rendering it neutral. No longer charged, the aspartate continues to move past the arginine.
Meanwhile, another aspartate residue encounters the continuation of the proton channel that leads onward to the matrix. It releases its proton, becoming charged. It is then attracted to the arginine in the stator, continuing the rotation of the rotor.
Exercise \(\PageIndex{2}\)
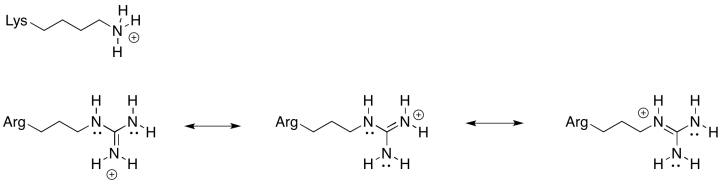
You could also imagine a lysine residue at the stator rather than the arginine shown above. Why is an arginine a more reliable positive stator than lysine?
- Answer
-
There is always an equilibrium between the protonated state and the deprotonated state in a charged amino acid residue. For this position, an amino acid is needed that is more reliably in the protonated state; that is, the equilibrium lies more heavily to the protonated side of the equation. Because of the resonance-stabilised cation that results from protonation, arginine is much more likely to remain in a protonated state than lysine. That will make for a more efficient millwheel.
ADP and phosphate bind at the interface of two proteins along this assembly. The rotation apparently is involved in an approximation mechanism. During the conformational changes that result from the molecular rotation, the ADP and phosphate are brought close enough together that they are more likely to bind to each other, forming ATP.
Exercise \(\PageIndex{3}\)
Why might it be difficult to bring ADP and phosphate together without this mechanism?
- Answer
-
ADP and phosphate are both anions; they would repel normally each other. When bound in the active site, their charges are likely neutralized by complementary charges in the active site.
Exercise \(\PageIndex{4}\)
Formation of an ATP from ADP costs about 30 kJ/mol. Assuming you could capture all of the energy released by transferring two electrons from NADH to O2, how many units of ATP could be made?
- Answer
-
Assuming the reduction potentials are:
NAD+ + 2e- + 2 H+ → NADH Eored = -0.32 V
0.5 O2 + 2e- + 2 H+ → H2O Eored = 0.816 V
Then the potential difference for the reaction, ΔEo = 0.816 - (-0.32) V = 1.136 V.
The Faraday relation ΔG = - n F ΔEo gives
ΔG = - 2 x 96,485 J V-1 mol-1 x 1.136 V = 219,213 J mol-1 = 219 kJ mol-1
so 219 kJ mol-1 / 30 kJ mol-1 = 7.3
With 100% efficiency, 7 moles of ATP could be produced per mole of NADH. In reality, about half that amount is produced (closer to 3 moles ATP per mole NADH).
See the focus page on the structure of Complex V at Biochemistry Online.
X-ray structures from: Watt, I.N., Montgomery, M.G., Runswick, M.J., Leslie, A.G.W., Walker, J.E. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc.Natl.Acad.Sci.USA 2010, 107, 16823. Images from RCSB Protein Data Bank (2XND).


