Image J Processing
- Page ID
- 242143
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \) \( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)\(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\) \(\newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\) \( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\) \( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\) \( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\) \( \newcommand{\Span}{\mathrm{span}}\)\(\newcommand{\AA}{\unicode[.8,0]{x212B}}\)
The following procedure outlines a standard approach to using Image J with TEM images. It is recommended that images be saved in .TIFF format.
Additional information, including examples and tutorials, can be found on the ImageJ homepage (https://imagej.nih.gov/ij/) in the Documentation link.
- Double click on the ImageJ shortcut on the desktop to open the image processing software.
- Load the TEM image from the desktop that corresponds to the synthesis conditions used by your lab group. Do this by selecting “File” and then “Open”.
- The software from modern TEM instruments will embed a scale bar on the image in order to give a reference to the size of objects in the image. We will use this to tell the software how big each pixel in the digital image is, which the software can then use to calculate sizes in a more automated manner.
- Use the zoom tool (looks like a magnifying glass) or the + and – keys on the keyboard to enlarge the image until the scale bar in the image almost fills the screen. Pan around the image using the tool that looks like a hand.
- Select the line tool from the tool bar at the top of the screen and draw a line over the scale bar (the line should extend exactly the same length as the scale bar).
- Select “Analyze” and then “Set Scale”.
- Remove any prior measurements by selecting “Click to remove scale”.
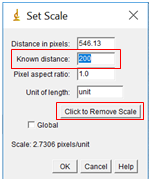
- In the box labelled “Known distance”, enter in the value on the scale bar in your image. The distance in pixels should already be filled from the line drawn. In the units box, enter the appropriate units, in this case nanometers or nm. Leave the pixel aspect ratio as the default value of 1 (this just means that the horizontal and vertical sizes of the pixels are the same). Click “OK”.
- Next be sure to zoom back out until you can see the entire image (hold control while clicking with the magnifying glass to zoom out), and then select the region of interest (where you have nanoparticles, but excluding the margins and scale bar). Do this by clicking on the rectangle tool and surrounding the area you want to use.
- Select “Image” and then “Crop”.
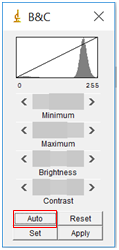
- You next need to adjust the image so that the software can readily distinguish between a nanoparticle and the background. To do this, you will first adjust the brightness/contrast of the image. Select “Image” followed by “Adjust” and then “Brightness/Contrast”. Sometimes the “Auto” feature is sufficient, but if not you can also play around with the sliding bars. Try to adjust them so that the nanoparticles really stand out and the background is mainly white, but make sure the size of the dots doesn’t change.
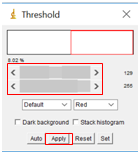
- Select “Image” followed by “Adjust” and then “Threshold”. Move the sliders until the particles you wish to analyze are red, but none of the background is red. This will set the background to be completely white to create a firm visual boundary between the nanoparticle and the background. Click “Apply”.
- Select “Process”, followed by “Binary” and then “Make Binary”.
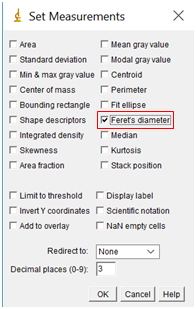
- We are about to measure the size of the particles, but first, we need to tell the software what measurements we want. Go to “Analyze” followed by “Set Measurements”. Check the box “Feret's Diameter”, but none of the others.
- Finally, we are ready to have the software measure the size of all the nanoparticles. To do this, select “Analyze” followed by “Analyze Particles”.
- Check the boxes “Display Results” and “Exclude on Edges” and “Summarize”. The default size from 0 to infinity is a good starting point, although it can be beneficial to put an upper and lower limit to prevent errant inclusions. For circularity, a range of 0.6 to 1 is typically acceptable.

- Select OK, and two tables with your results will appear, a summary and complete table of each particle.
- Look through the results table and watch for unrealistic particle sizes (for example if the area is on the order of 1x10-4 nm2, this makes no physical sense, it results from background noise being counted as a particle). You can remove these errant points by right clicking on them and selecting “clear”.
- For each window, select “File” followed by “save as” to save the table as a spreadsheet. End your filename with “.txt”. For the summary table, either print the spreadsheet or copy the results into your lab notebook.
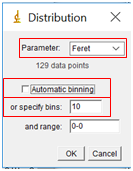
- While the complete data table is nice, it is challenging to visualize the results in table form. Instead, we will create a histogram which shows the size distribution of our particles. In the Results window, select “Results” followed by “Set Measurements”. Select the Feret’s Diameter box (you can uncheck all the others). Then select “Results” followed by “Distribution”. Under “Parameter”, select “Feret”. Uncheck “Automatic Binning” and set the number of bins to 10, and then select “OK”. Note that the binning determines how many bars are displayed on the histogram.
- Save your data for future use. You can also copy data (edit>select all>copy) and paste in Excel or comparable software to analyze the data and create graphs.


